The Potential of Digested Sludge-Assimilating Microflora for Biogas Production from Food Processing Wastes
- PMID: 37764166
- PMCID: PMC10535770
- DOI: 10.3390/microorganisms11092321
The Potential of Digested Sludge-Assimilating Microflora for Biogas Production from Food Processing Wastes
Abstract
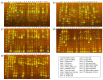
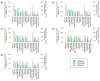
Food processing wastes (FPWs) are residues generated in food manufacturing, and their composition varies depending on the type of food product being manufactured. Therefore, selecting and acclimatizing seed microflora during the initiation of biogas production is crucial for optimal outcomes. The present study examined the biogas production capabilities of digested sludge-assimilating and biogas-yielding soil (DABYS) and enteric (DABYE) microflorae when used as seed cultures for biogas production from FPWs. After subculturing and feeding these microbial seeds with various FPWs, we assessed their biogas-producing abilities. The subcultures produced biogas from many FPWs, except orange peel, suggesting that the heterogeneity of the bacterial members in the seed microflora facilitates quick adaptation to FPWs. Microflorae fed with animal-derived FPWs contained several methanogenic archaeal families and produced methane. In contrast, microflorae fed with vegetable-, fruit-, and crop-derived FPWs generated hydrogen, and methanogenic archaeal populations were diminished by repeated subculturing. The subcultured microflorae appear to hydrolyze carbohydrates and protein in FPWs using cellulase, pectinase, or protease. Despite needing enhancements in biogas yield for future industrial scale-up, the DABYS and DABYE microflorae demonstrate robust adaptability to various FPWs.
Keywords: anaerobic digestion; cellulase; food waste recycling; hydrogen; methane; pectinase; protease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- UNEP . Food Waste Index Report 2021. UNEP; Nairobi, Kenya: 2021. [(accessed on 25 July 2023)]. Food waste amounts: Measured estimates and extrapolations; pp. 54–72. Available online: https://www.unep.org/resources/report/unep-food-waste-index-report-2021.
-
- FAO . The State of Food and Agriculture. FAO; Rome, Italy: 2019. [(accessed on 25 July 2023)]. Food Loss and Waste—Framing the Issues; pp. 1–19. Available online: https://www.fao.org/3/ca6030en/ca6030en.pdf.
-
- Zhang C., Kang X., Wang F., Tian Y., Liu T., Su Y., Qian T., Zhang Y. Valorization of food waste for cost-effective reducing sugar recovery in a two-stage enzymatic hydrolysis platform. Energy. 2020;208:118379. doi: 10.1016/j.energy.2020.118379. - DOI
-
- Ali A., Bux Mahar R., Panhwar S., Ahmed Keerio H., Hussain Khokhar N., Suja F., Rundong L. Generation of green renewable energy through anaerobic digestion technology (ADT): Technical insights review. Waste Biomass Valoriz. 2023;14:663–686. doi: 10.1007/s12649-022-02001-7. - DOI
-
- Ferdeș M., Zăbavă B.Ș., Paraschiv G., Ionescu M., Dincă M.N., Moiceanu G. Food waste management for biogas production in the context of sustainable development. Energies. 2022;15:6268. doi: 10.3390/en15176268. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

