Einstein Model of a Graph to Characterize Protein Folded/Unfolded States
- PMID: 37764437
- PMCID: PMC10536427
- DOI: 10.3390/molecules28186659
Einstein Model of a Graph to Characterize Protein Folded/Unfolded States
Abstract
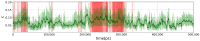
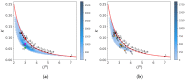
The folded structures of proteins can be accurately predicted by deep learning algorithms from their amino-acid sequences. By contrast, in spite of decades of research studies, the prediction of folding pathways and the unfolded and misfolded states of proteins, which are intimately related to diseases, remains challenging. A two-state (folded/unfolded) description of protein folding dynamics hides the complexity of the unfolded and misfolded microstates. Here, we focus on the development of simplified order parameters to decipher the complexity of disordered protein structures. First, we show that any connected, undirected, and simple graph can be associated with a linear chain of atoms in thermal equilibrium. This analogy provides an interpretation of the usual topological descriptors of a graph, namely the Kirchhoff index and Randić resistance, in terms of effective force constants of a linear chain. We derive an exact relation between the Kirchhoff index and the average shortest path length for a linear graph and define the free energies of a graph using an Einstein model. Second, we represent the three-dimensional protein structures by connected, undirected, and simple graphs. As a proof of concept, we compute the topological descriptors and the graph free energies for an all-atom molecular dynamics trajectory of folding/unfolding events of the proteins Trp-cage and HP-36 and for the ensemble of experimental NMR models of Trp-cage. The present work shows that the local, nonlocal, and global force constants and free energies of a graph are promising tools to quantify unfolded/disordered protein states and folding/unfolding dynamics. In particular, they allow the detection of transient misfolded rigid states.
Keywords: Kirchhoff index; Wiener index; graph theory; intrinsically disordered proteins; molecular dynamics; protein folding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Unfolded protein ensembles, folding trajectories, and refolding rate prediction.J Chem Phys. 2013 Sep 28;139(12):121925. doi: 10.1063/1.4817215. J Chem Phys. 2013. PMID: 24089737
-
Protonation/deprotonation effects on the stability of the Trp-cage miniprotein.Phys Chem Chem Phys. 2011 Oct 14;13(38):17056-63. doi: 10.1039/c1cp21193e. Epub 2011 Jul 20. Phys Chem Chem Phys. 2011. PMID: 21773639
-
Comparison of hydration behavior and conformational preferences of the Trp-cage mini-protein in different rigid-body water models.Phys Chem Chem Phys. 2016 Dec 7;18(48):32796-32813. doi: 10.1039/c6cp04634g. Phys Chem Chem Phys. 2016. PMID: 27878168
-
Limited cooperativity in protein folding.Curr Opin Struct Biol. 2016 Feb;36:58-66. doi: 10.1016/j.sbi.2015.12.001. Epub 2016 Feb 2. Curr Opin Struct Biol. 2016. PMID: 26845039 Review.
-
Dynamics of protein folding: probing the kinetic network of folding-unfolding transitions with experiment and theory.Biochim Biophys Acta. 2011 Aug;1814(8):1001-20. doi: 10.1016/j.bbapap.2010.09.013. Epub 2010 Sep 29. Biochim Biophys Acta. 2011. PMID: 20883829 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

