Comparative Analysis of Campylobacter jejuni and C. coli Isolated from Livestock Animals to C. jejuni and C. coli Isolated from Surface Water Using DNA Sequencing and MALDI-TOF
- PMID: 37764877
- PMCID: PMC10535298
- DOI: 10.3390/pathogens12091069
Comparative Analysis of Campylobacter jejuni and C. coli Isolated from Livestock Animals to C. jejuni and C. coli Isolated from Surface Water Using DNA Sequencing and MALDI-TOF
Abstract
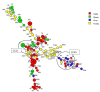
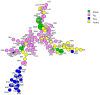
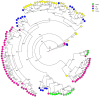
This study evaluated the contribution of cattle, sheep, poultry and pigs to the contamination of surface water from rivers by Campylobacter jejuni and C. coli using MLST, cgMLST and considered MALDI-TOF MS as an alternative technique. The 263 strains isolated from cattle (n = 61), sheep (n = 42), poultry (n = 65), pigs (n = 60) and surface water (n = 35) were distributed across 115 sequence types (STs), 49 for C. jejuni and 66 for C. coli. Considering MLST data, 14.2%, 11.4% and 2.8% of the surface water strains could be attributed to cattle, poultry and sheep, respectively, none to pigs, and 85.7% were non-attributed. Analysis of cg-MLST data with STRUCTURE indicated that C. jejuni strains from water were predominantly attributed to poultry (93.5%), weakly to sheep (<1%) and 6.3% non-attributed, and that conversely, C. coli strains from water were predominantly non-attributed (94.3%) and 5.7% attributed to poultry. Considering the protein profiles with a threshold of 94% and 97% of similarity, respectively, strains from surface water could be attributed to poultry (31.4% and 17.1%), and to cattle (17.1% and 5.7%); 54.1% and 77.1% were non-attributed. This study confirmed these livestock animals might contribute to the contamination of surface water, with a level of contribution depending on the typing technique and the method of analysis. MALDI-TOF could potentially be an alternative approach for source attribution.
Keywords: Campylobacter; MALDI-TOF; MLST; cgMLST; livestock animals; structure; surface water.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Tracing the animal sources of surface water contamination with Campylobacter jejuni and Campylobacter coli.Water Res. 2020 Dec 15;187:116421. doi: 10.1016/j.watres.2020.116421. Epub 2020 Sep 20. Water Res. 2020. PMID: 32992147
-
Species Identification of Campylobacter jejuni and Campylobacter coli Isolates from Raw Poultry Products by MALDI-TOF MS and rRNA Sequence Analysis.J AOAC Int. 2020 Jan 1;103(1):197-204. doi: 10.5740/jaoacint.19-0170. J AOAC Int. 2020. PMID: 31324272
-
A pilot study revealing host-associated genetic signatures for source attribution of sporadic Campylobacter jejuni infection in Egypt.Transbound Emerg Dis. 2022 Jul;69(4):1847-1861. doi: 10.1111/tbed.14165. Epub 2021 Jun 9. Transbound Emerg Dis. 2022. PMID: 34033263
-
Genetic diversity among Campylobacter jejuni isolates from healthy livestock and their links to human isolates in Spain.Zoonoses Public Health. 2011 Aug;58(5):365-75. doi: 10.1111/j.1863-2378.2010.01373.x. Epub 2010 Oct 7. Zoonoses Public Health. 2011. PMID: 21040505
-
Prevalence and diversity of Campylobacter jejuni in pig herds on farms with and without cattle or poultry.J Food Prot. 2005 Apr;68(4):722-7. doi: 10.4315/0362-028x-68.4.722. J Food Prot. 2005. PMID: 15830662
Cited by
-
Antimicrobial Effects of Plant-Based Supplements on Gut Microbial Diversity in Small Ruminants.Pathogens. 2023 Dec 29;13(1):31. doi: 10.3390/pathogens13010031. Pathogens. 2023. PMID: 38251338 Free PMC article. Review.
References
-
- Van Cauteren D., De Valk H., Sommen C., King L.A., Jourdan-Da Silva N., Weill F.X., Le Hello S., Mégraud F., Vaillant V., Desenclos J.C. Community Incidence of Campylobacteriosis and Nontyphoidal Salmonellosis, France, 2008–2013. Foodborne Pathog. Dis. 2015;12:664–669. doi: 10.1089/fpd.2015.1964. - DOI - PubMed
-
- CNCH (Centre National de Référence des Campylobacters et des Helicobacters) SPF (Santé Publique France) Rapport de l’Année d’Exercice 2019. CNCH; Paris, France: 2020. 64p CNCH Report.
-
- Thépault A., Méric G., Rivoal K., Pascoe B., Mageiros L., Touzain F., Rose V., Béven V., Chemaly M., Sheppard S.K. Genome-wide identification of host-segregating epidemiological markers for source attribution in Campylobacter jejuni. Appl. Environ. Microbiol. 2017;83:e03085–e030816. doi: 10.1128/AEM.03085-16. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

