Porcine Respiratory Coronavirus (PRCV): Isolation and Characterization of a Variant PRCV from USA Pigs
- PMID: 37764905
- PMCID: PMC10536027
- DOI: 10.3390/pathogens12091097
Porcine Respiratory Coronavirus (PRCV): Isolation and Characterization of a Variant PRCV from USA Pigs
Abstract
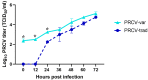
Porcine respiratory coronavirus (PRCV), a mutant of the transmissible gastroenteritis virus (TGEV), was first reported in Belgium in 1984. PRCV typically replicates and induces mild lesions in the respiratory tract, distinct from the enteric tropism of TGEV. In the past 30 years, PRCV has rarely been studied, and most cited information is on traditional isolates obtained during the 1980s and 1990s. Little is known about the genetic makeup and pathogenicity of recent PRCV isolates. The objective of this study was to obtain a contemporary PRCV isolate from US pigs for genetic characterization. In total, 1245 lung homogenate samples from pigs in various US states were tested via real-time PCR targeting PRCV and TGEV RNA. Overall, PRCV RNA was detected in five samples, and a single isolate (ISU20-92330) was successfully cultured and sequenced for its full-length genome. The isolate clustered with a new group of variant TGEVs and differed in various genomic regions compared to traditional PRCV isolates. Pathogens, such as PRCV, commonly circulate in pig herds without causing major disease. There may be value in tracking genomic changes and regularly updating the diagnostic methods for such viruses to be better prepared for the emergence of variants in ecology and pathogenicity.
Keywords: PRCV/TGEV/XIPC 3-plex RT PCR; genomic analysis; porcine respiratory coronavirus (PRCV); prevalence; transmissible gastroenteritis virus (TGEV).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

