HSV-1 Triggers an Antiviral Transcriptional Response during Viral Replication That Is Completely Abrogated in PKR-/- Cells
- PMID: 37764935
- PMCID: PMC10536113
- DOI: 10.3390/pathogens12091126
HSV-1 Triggers an Antiviral Transcriptional Response during Viral Replication That Is Completely Abrogated in PKR-/- Cells
Abstract
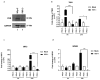
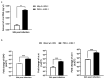
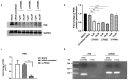
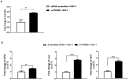
The activation of the innate immune response during HSV-1 infection stimulates several transcription factors, such as NF-κB and IRF3, which are critical regulators of IFN-β expression. The released IFN-β activates the ISGs, which encode antiviral effectors such as the PKR. We found that HSV-1 triggers an antiviral transcriptional response during viral replication by activating TBK1-IRF3-NF-κB network kinetically. In contrast, we reported that infected PKR-/- cells fail to activate the transcription of TBK1. Downstream, TBK1 was unable to activate the transcription of IRF3 and NF-κB. These data suggested that in PKR-/- cells, HSV-1 replication counteracts TBK1-IRF3-NF-κB network. In this scenario, a combined approach of gene knockout and gene silencing was used to determine how the lack of PKR facilitates HSV-1 replication. We reported that in HEp-2-infected cells, PKR can influence the TBK1-IRF3-NF-κB network, consequently interfering with viral replication. Otherwise, an abrogated PKR-mediated signaling sustains the HSV-1 replication. Our result allows us to add additional information on the complex HSV-host interaction network by reinforcing the concept of the PKR role in the innate response-related networks during HSV replication in an in vitro model.
Keywords: HSV-1; PKR; innate antiviral response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The early interferon response to rotavirus is regulated by PKR and depends on MAVS/IPS-1, RIG-I, MDA-5, and IRF3.J Virol. 2011 Apr;85(8):3717-32. doi: 10.1128/JVI.02634-10. Epub 2011 Feb 9. J Virol. 2011. PMID: 21307186 Free PMC article.
-
Herpes Simplex Virus 1 Tegument Protein UL46 Inhibits TANK-Binding Kinase 1-Mediated Signaling.mBio. 2019 May 21;10(3):e00919-19. doi: 10.1128/mBio.00919-19. mBio. 2019. PMID: 31113902 Free PMC article.
-
Heartland virus NSs protein disrupts host defenses by blocking the TBK1 kinase-IRF3 transcription factor interaction and signaling required for interferon induction.J Biol Chem. 2017 Oct 6;292(40):16722-16733. doi: 10.1074/jbc.M117.805127. Epub 2017 Aug 28. J Biol Chem. 2017. PMID: 28848048 Free PMC article.
-
The Molecular Mechanism of Herpes Simplex Virus 1 UL31 in Antagonizing the Activity of IFN-β.Microbiol Spectr. 2022 Feb 23;10(1):e0188321. doi: 10.1128/spectrum.01883-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196784 Free PMC article.
-
TANK-binding kinase 1 as a novel therapeutic target for viral diseases.Expert Opin Ther Targets. 2019 May;23(5):437-446. doi: 10.1080/14728222.2019.1601702. Epub 2019 Apr 10. Expert Opin Ther Targets. 2019. PMID: 30932713 Review.
Cited by
-
Understanding and therapeutically exploiting cGAS/STING signaling in glioblastoma.J Clin Invest. 2024 Jan 16;134(2):e163452. doi: 10.1172/JCI163452. J Clin Invest. 2024. PMID: 38226619 Free PMC article. Review.
-
Post-translational modifications as a key mechanism for herpes simplex virus type I evasion of host innate immunity.Front Microbiol. 2025 Feb 11;16:1543676. doi: 10.3389/fmicb.2025.1543676. eCollection 2025. Front Microbiol. 2025. PMID: 40008039 Free PMC article. Review.
-
How Does African Swine Fever Virus Evade the cGAS-STING Pathway?Pathogens. 2024 Nov 2;13(11):957. doi: 10.3390/pathogens13110957. Pathogens. 2024. PMID: 39599510 Free PMC article. Review.
-
ADAR1 p150 prevents HSV-1 from triggering PKR/eIF2α-mediated translational arrest and is required for efficient viral replication.PLoS Pathog. 2025 Apr 8;21(4):e1012452. doi: 10.1371/journal.ppat.1012452. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40198737 Free PMC article.
References
-
- Stelitano D., Franci G., Chianese A., Galdiero S., Morelli G., Galdiero M. HSV membrane glycoproteins, their function in viral entry and their use in vaccine studies. In: Ryadnov M., Hudecz F., editors. Amino Acids, Peptides and Proteins. Volume 43. The Royal Society of Chemistry; Cambridge, UK: 2019. pp. 14–43.
LinkOut - more resources
Full Text Sources
Miscellaneous

