Genome-Wide Identification and Analysis of TCP Gene Family among Three Dendrobium Species
- PMID: 37765364
- PMCID: PMC10538224
- DOI: 10.3390/plants12183201
Genome-Wide Identification and Analysis of TCP Gene Family among Three Dendrobium Species
Abstract
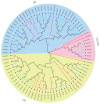
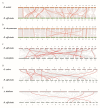
Dendrobium orchids, which are among the most well-known species of orchids, are appreciated for their aesthetic appeal across the globe. Furthermore, due to their strict living conditions, they have accumulated high levels of active ingredients, resulting not only in their medicinal value but also in their strong ability to respond to harsh environments. The TCP gene family plays an important role in plant growth and development, and signal transduction. However, these genes have not been systematically investigated in Dendrobium species. In this study, we detected a total of 24, 23, and 14 candidate TCP members in the genome sequences of D. officinale, D. nobile, and D. chrysotoxum, respectively. These genes were classified into three clades on the basis of a phylogenetic analysis. The TCP gene numbers among Dendrobium species were still highly variable due to the independent loss of genes in the CIN clade. However, only three gene duplication events were detected, with only one tandem duplication event (DcTCP9/DcTCP10) in D. chrysotoxum and two pairs of paralogous DoTCP gene duplication events (DoTCP1/DoTCP23 and DoTCP16/DoTCP24) in D. officinale. A total of 25 cis-acting elements of TCPs related to hormone/stress and light responses were detected. Among them, the proportions of hormone response, light response, and stress response elements in D. officinale (100/421, 127/421, and 171/421) were similar to those in D. nobile (83/352, 87/352, and 161/352). Using qRT-PCR to determine their expression patterns under MeJA treatment, four DoTCPs (DoTCP2, DoTCP4, DoTCP6, and DoTCP14) were significantly upregulated under MeJA treatment, which indicates that TCP genes may play important roles in responding to stress. Under ABA treatment, seven DoTCPs (DoTCP3, DoTCP7, DoTCP9, DoTCP11, DoTCP14, DoTCP15, and DoTCP21) were significantly upregulated, indicating that TCP genes may also play an important role in hormone response. Therefore, these results can provide useful information for studying the evolution and function of TCP genes in Dendrobium species.
Keywords: Dendrobium; TCP proteins; expression profiles; gene family; genome-wide analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Genome-wide identification and expression analysis of growth-regulating factors in Dendrobium officinale and Dendrobium chrysotoxum.PeerJ. 2023 Dec 15;11:e16644. doi: 10.7717/peerj.16644. eCollection 2023. PeerJ. 2023. PMID: 38111654 Free PMC article.
-
Genome-Wide Identification of TCP Gene Family in Dendrobium and Their Expression Patterns in Dendrobium chrysotoxum.Int J Mol Sci. 2023 Sep 20;24(18):14320. doi: 10.3390/ijms241814320. Int J Mol Sci. 2023. PMID: 37762622 Free PMC article.
-
Genome-wide identification and characterization of TCP gene family in Dendrobium nobile and their role in perianth development.Front Plant Sci. 2024 Feb 5;15:1352119. doi: 10.3389/fpls.2024.1352119. eCollection 2024. Front Plant Sci. 2024. PMID: 38375086 Free PMC article.
-
Genome-Wide Identification of the YABBY Gene Family in Dendrobium Orchids and Its Expression Patterns in Dendrobium chrysotoxum.Int J Mol Sci. 2023 Jun 15;24(12):10165. doi: 10.3390/ijms241210165. Int J Mol Sci. 2023. PMID: 37373311 Free PMC article.
-
Genome-wide identification of Ankyrin (ANK) repeat gene families in three Dendrobium species and the expression of ANK genes in D. officinale under gibberellin and abscisic acid treatments.BMC Plant Biol. 2024 Aug 10;24(1):762. doi: 10.1186/s12870-024-05461-2. BMC Plant Biol. 2024. PMID: 39123107 Free PMC article.
Cited by
-
Genome-wide identification and stress-responsive expression profiling of the TCP gene family in Cenchrus fungigraminus under drought and cold stress.BMC Genomics. 2025 Jul 1;26(1):626. doi: 10.1186/s12864-025-11798-1. BMC Genomics. 2025. PMID: 40596840 Free PMC article.
-
Evolution and comparison of the expression of TCP genes in the benincaseae and cucurbiteae tribes.Sci Rep. 2025 May 3;15(1):15470. doi: 10.1038/s41598-025-99296-y. Sci Rep. 2025. PMID: 40316658 Free PMC article.
-
Genome-wide identification and expression analysis of the TCP transcription factor family and its response to abiotic stress in rapeseed (Brassica napus L.).3 Biotech. 2025 May;15(5):119. doi: 10.1007/s13205-025-04273-x. Epub 2025 Apr 7. 3 Biotech. 2025. PMID: 40201755 Free PMC article.
References
-
- Chen P., Li J., Ye X., Tan B., Zheng Z., Cheng J., Wang W., Wang H., Gu L., Feng J., et al. Genome-wide identification of Ziziphus jujuba TCP transcription factors and their expression in response to infection with jujube witches’ broom phytoplasma. Acta Physiol. Plant. 2019;41:86. doi: 10.1007/s11738-019-2879-9. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

