Site-Directed Mutations at Phosphorylation Sites in Zea mays PHO1 Reveal Modulation of Enzymatic Activity by Phosphorylation at S566 in the L80 Region
- PMID: 37765369
- PMCID: PMC10536461
- DOI: 10.3390/plants12183205
Site-Directed Mutations at Phosphorylation Sites in Zea mays PHO1 Reveal Modulation of Enzymatic Activity by Phosphorylation at S566 in the L80 Region
Abstract
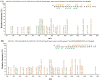
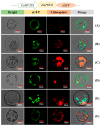
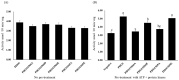
Starch phosphorylase (PHO) is a pivotal enzyme within the GT35-glycogen-phosphorylase (GT; glycosyltransferases) superfamily. Despite the ongoing debate surrounding the precise role of PHO1, evidence points to its substantial influence on starch biosynthesis, supported by its gene expression profile and subcellular localization. Key to PHO1 function is the enzymatic regulation via phosphorylation; a myriad of such modification sites has been unveiled in model crops. However, the functional implications of these sites remain to be elucidated. In this study, we utilized site-directed mutagenesis on the phosphorylation sites of Zea mays PHO1, replacing serine residues with alanine, glutamic acid, and aspartic acid, to discern the effects of phosphorylation. Our findings indicate that phosphorylation exerts no impact on the stability or localization of PHO1. Nonetheless, our enzymatic assays unveiled a crucial role for phosphorylation at the S566 residue within the L80 region of the PHO1 structure, suggesting a potential modulation or enhancement of PHO1 activity. These data advance our understanding of starch biosynthesis regulation and present potential targets for crop yield optimization.
Keywords: activity; phosphorylation; plastidial starch phosphorylase; subcellular localization.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Serine 31 Phosphorylation-Driven Regulation of AGPase Activity: Potential Implications for Enhanced Starch Yields in Crops.Int J Mol Sci. 2023 Oct 18;24(20):15283. doi: 10.3390/ijms242015283. Int J Mol Sci. 2023. PMID: 37894964 Free PMC article.
-
Molecular Functions and Pathways of Plastidial Starch Phosphorylase (PHO1) in Starch Metabolism: Current and Future Perspectives.Int J Mol Sci. 2021 Sep 28;22(19):10450. doi: 10.3390/ijms221910450. Int J Mol Sci. 2021. PMID: 34638789 Free PMC article. Review.
-
Comparative Study of Starch Phosphorylase Genes and Encoded Proteins in Various Monocots and Dicots with Emphasis on Maize.Int J Mol Sci. 2022 Apr 20;23(9):4518. doi: 10.3390/ijms23094518. Int J Mol Sci. 2022. PMID: 35562912 Free PMC article.
-
The plastid phosphorylase as a multiple-role player in plant metabolism.Plant Sci. 2020 Jan;290:110303. doi: 10.1016/j.plantsci.2019.110303. Epub 2019 Oct 10. Plant Sci. 2020. PMID: 31779913 Review.
-
The Rice Plastidial Phosphorylase Participates Directly in Both Sink and Source Processes.Plant Cell Physiol. 2021 Mar 25;62(1):125-142. doi: 10.1093/pcp/pcaa146. Plant Cell Physiol. 2021. PMID: 33237266
Cited by
-
Cloning and function analysis of ZmICE1a, a contributor to the melioration of maize kernel traits.Plant Signal Behav. 2025 Dec;20(1):2521320. doi: 10.1080/15592324.2025.2521320. Epub 2025 Jul 2. Plant Signal Behav. 2025. PMID: 40600923 Free PMC article.
-
Serine 31 Phosphorylation-Driven Regulation of AGPase Activity: Potential Implications for Enhanced Starch Yields in Crops.Int J Mol Sci. 2023 Oct 18;24(20):15283. doi: 10.3390/ijms242015283. Int J Mol Sci. 2023. PMID: 37894964 Free PMC article.
References
-
- Cori G.T., Cori C.F., Schmidt G. The role of glucose-l-phosphate in the formation of blood sugar and synthesis of glycogen in the liver. J. Biol. Chem. 1939;142:355–366. doi: 10.1016/S0021-9258(18)73628-4. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

