Investigating Variability in Viral Presence and Abundance across Soybean Seed Development Stages Using Transcriptome Analysis
- PMID: 37765420
- PMCID: PMC10535271
- DOI: 10.3390/plants12183257
Investigating Variability in Viral Presence and Abundance across Soybean Seed Development Stages Using Transcriptome Analysis
Abstract
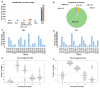
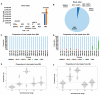
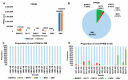
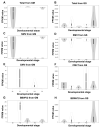
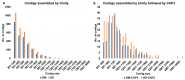
Plant transcriptomes offer a valuable resource for studying viral communities (viromes). In this study, we explore how plant transcriptome data can be applied to virome research. We analyzed 40 soybean transcriptomes across different growth stages and identified six viruses: broad bean wilt virus 2 (BBWV2), brassica yellow virus (BrYV), beet western yellow virus (BWYV), cucumber mosaic virus (CMV), milk vetch dwarf virus (MDV), and soybean mosaic virus (SMV). SMV was the predominant virus in both Glycine max (GM) and Glycine soja (GS) cultivars. Our analysis confirmed its abundance in both, while BBWV2 and CMV were more prevalent in GS than GM. The viral proportions varied across developmental stages, peaking in open flowers. Comparing viral abundance measured by viral reads and fragments per kilobase of transcript per million (FPKM) values revealed insights. SMV showed similar FPKM values in GM and GS, but BBWV2 and CMV displayed higher FPKM proportions in GS. Notably, the differences in viral abundance between GM and GS were generally insignificant based on the FPKM values across developmental stages, except for the apical bud stage in four GM cultivars. We also detected MDV, a multi-segmented virus, in two GM samples, with variable proportions of its segments. In conclusion, our study demonstrates the potential of plant transcriptomes for virome research, highlighting their strengths and limitations.
Keywords: soybean seed development; transcriptome analysis; viral load shifts; viromes; virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Toomer O.T., Oviedo E.O., Ali M., Patino D., Joseph M., Frinsko M., Vu T., Maharjan P., Fallen B., Mian R. Current agronomic practices, harvest & post-harvest processing of soybeans (Glycine max)—A review. Agronomy. 2023;13:427.
-
- Hill J.H., Whitham S.A. Advances in Virus Research. Volume 90. Elsevier; Amsterdam, The Netherlands: 2014. Control of virus diseases in soybeans; pp. 355–390. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

