Phenotypic Variation Analysis and Excellent Clone Selection of Alnus cremastogyne from Different Provenances
- PMID: 37765423
- PMCID: PMC10535346
- DOI: 10.3390/plants12183259
Phenotypic Variation Analysis and Excellent Clone Selection of Alnus cremastogyne from Different Provenances
Abstract
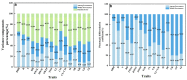
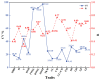
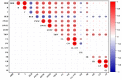
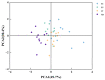
Alnus cremastogyne is a rapidly growing broad-leaved tree species that is widely distributed in southwest China. It has a significant economic and ecological value. However, with the expansion of the planting area, the influence of phenotypic variation and differentiation on Alnus cremastogyne has increased, resulting in a continuous decline in its genetic quality. Therefore, it is crucial to investigate the phenotypic variation of Alnus cremastogyne and select excellent breeding materials for genetic improvement. Herein, four growth-related phenotypic traits (diameter at breast height, the height of trees, volume, height under the branches) and twelve reproductive-related phenotypic traits (fresh weight of single cone, dry weight of single cone, seed weight per plant, thousand kernel weight, cone length, cone width, cone length × cone width, fruit shape index, seed rate, germination rate, germination potential, germination index) of 40 clones from four provenances were measured and analyzed. The phenotypic variation was comprehensively evaluated by correlation analysis, principal component analysis and cluster analysis, and excellent clones were selected as breeding materials. The results revealed that there were abundant phenotypic traits variations among and within provenances. Most of the phenotypic traits were highly significant differences (p < 0.01) among provenances. The phenotypic variation among provenances (26.36%) was greater than that of within provenances clones (24.80%). The average phenotypic differentiation coefficient was accounted for 52.61% among provenances, indicating that the phenotypic variation mainly came from among provenances. The coefficient of variation ranged from 9.41% (fruit shape index) to 97.19% (seed weight per plant), and the repeatability ranged from 0.36 (volume) to 0.77 (cone width). Correlation analysis revealed a significantly positive correlation among most phenotypic traits. In principal component analysis, the cumulative contribution rate of the first three principal components was 79.18%, representing the main information on the measured phenotypic traits. The cluster analysis revealed four groups for the 40 clones. Group I and group II exhibited better performance phenotypic traits as compared with group III and group IV. In addition, the four groups are not clearly clustered following the distance from the provenance. Employing the multi-trait comprehensive evaluation method, 12 excellent clones were selected, and the average genetic gain for each phenotypic trait ranged from 4.78% (diameter at breast height) to 32.05% (dry weight of single cone). These selected excellent clones can serve as candidate materials for the improvement and transformation of Alnus cremastogyne seed orchards. In addition, this study can also provide a theoretical foundation for the genetic improvement, breeding, and clone selection of Alnus cremastogyne.
Keywords: Alnus cremastogyne; excellent clone; phenotypic traits; phenotypic variation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Variations in Growth and Photosynthetic Traits of Polyploid Poplar Hybrids and Clones in Northeast China.Genes (Basel). 2022 Nov 19;13(11):2161. doi: 10.3390/genes13112161. Genes (Basel). 2022. PMID: 36421836 Free PMC article.
-
Provenance and family variations in early growth of Manchurian walnut (Juglans mandshurica Maxim.) and selection of superior families.PLoS One. 2024 Mar 7;19(3):e0298918. doi: 10.1371/journal.pone.0298918. eCollection 2024. PLoS One. 2024. PMID: 38451964 Free PMC article.
-
Analysis and evaluation of Camellia oleifera Abel. Germplasm fruit traits from the high-altitude areas of East Guizhou Province, China.Sci Rep. 2024 Aug 8;14(1):18440. doi: 10.1038/s41598-024-69454-9. Sci Rep. 2024. PMID: 39117844 Free PMC article.
-
Analysis of Components and Properties of Extractives from Alnus cremastogyne Pods from Different Provenances.Molecules. 2022 Nov 12;27(22):7802. doi: 10.3390/molecules27227802. Molecules. 2022. PMID: 36431903 Free PMC article.
-
Variation of phenotypic and physiological traits of Robinia pseudoacacia L. from 20 provenances.PLoS One. 2022 Jan 5;17(1):e0262278. doi: 10.1371/journal.pone.0262278. eCollection 2022. PLoS One. 2022. PMID: 34986177 Free PMC article.
Cited by
-
Differences in Physiological and Agronomic Traits and Evaluation of Adaptation of Seven Maize Varieties.Biology (Basel). 2024 Nov 26;13(12):977. doi: 10.3390/biology13120977. Biology (Basel). 2024. PMID: 39765645 Free PMC article.
-
Impact of Heavy Metal Pollution in the Environment on the Metabolic Profile of Medicinal Plants and Their Therapeutic Potential.Plants (Basel). 2024 Mar 21;13(6):913. doi: 10.3390/plants13060913. Plants (Basel). 2024. PMID: 38592933 Free PMC article. Review.
-
Phenotypic diversity and provenance variation of Cupressus funebris: a case study in the Sichuan Basin, China.PeerJ. 2024 Nov 29;12:e18494. doi: 10.7717/peerj.18494. eCollection 2024. PeerJ. 2024. PMID: 39624132 Free PMC article.
-
Screening for cold tolerance resources in maize seedlings and analysis of leaf cell responses.Front Plant Sci. 2025 Jun 25;16:1565831. doi: 10.3389/fpls.2025.1565831. eCollection 2025. Front Plant Sci. 2025. PMID: 40636019 Free PMC article.
-
Genetic Diversity Analysis of Water Lily Germplasms Based on Morphological Traits and SSR Markers.Plants (Basel). 2025 Apr 30;14(9):1365. doi: 10.3390/plants14091365. Plants (Basel). 2025. PMID: 40364393 Free PMC article.
References
-
- Rao L., Li Y., Guo H., Duan H., Chen Y. Comparisons on seedlings growth traits of five alder genus species. J. Cent. South Univ. For. Technol. 2016;36:18–25. doi: 10.14067/j.cnki.1673-923x.2016.01.004. (In Chinese) - DOI
-
- Guo H., Yang H., Chen Z., Wang Z., Huang Z., Li J., Xiao X., Kang X. Genetic Effects of Fruit, Seed and Seedling Traits of Alnus cremastogyne Burk in a 7 × 7 Complete Diallel Cross Design. Bull. Bot. Res. 2018;38:357–366. doi: 10.7525/j.issn.1673-5102.2018.03.007. (In Chinese) - DOI
-
- Chen B., Zhou Z., Li G., Huang G., Yang L. Status on Pulping/Papering Research of Alder and Prospect of Utilization of Alder Pulp Wood in China. For. Res. 1999;12:656–661. (In Chinese)
-
- Liu L., Wang R., Zhang Y., Mou Q., Gou Y., Liu K., Huang N., Ouyang C., Hu J., Du B. Simulation of potential suitable distribution of Alnus cremastogyne Burk. In China under climate change scenarios. Ecol. Indic. 2021;133:108396. doi: 10.1016/j.ecolind.2021.108396. - DOI
LinkOut - more resources
Full Text Sources

