Evolutionary Dynamics of FLC-like MADS-Box Genes in Brassicaceae
- PMID: 37765445
- PMCID: PMC10536770
- DOI: 10.3390/plants12183281
Evolutionary Dynamics of FLC-like MADS-Box Genes in Brassicaceae
Abstract
MADS-box genes encode transcription factors that play important roles in the development and evolution of plants. There are more than a dozen clades of MADS-box genes in angiosperms, of which those with functions in the specification of floral organ identity are especially well-known. From what has been elucidated in the model plant Arabidopsis thaliana, the clade of FLC-like MADS-box genes, comprising FLC-like genes sensu strictu and MAF-like genes, are somewhat special among the MADS-box genes of plants since FLC-like genes, especially MAF-like genes, show unusual evolutionary dynamics, in that they generate clusters of tandemly duplicated genes. Here, we make use of the latest genomic data of Brassicaceae to study this remarkable feature of the FLC-like genes in a phylogenetic context. We have identified all FLC-like genes in the genomes of 29 species of Brassicaceae and reconstructed the phylogeny of these genes employing a Maximum Likelihood method. In addition, we conducted selection analyses using PAML. Our results reveal that there are three major clades of FLC-like genes in Brassicaceae that all evolve under purifying selection but with remarkably different strengths. We confirm that the tandem arrangement of MAF-like genes in the genomes of Brassicaceae resulted in a high rate of duplications and losses. Interestingly, MAF-like genes also seem to be prone to transposition. Considering the role of FLC-like genes sensu lato (s.l.) in the timing of floral transition, we hypothesize that this rapid evolution of the MAF-like genes was a main contributor to the successful adaptation of Brassicaceae to different environments.
Keywords: MADS-box gene; comparative transcriptomics; phylogenomics; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
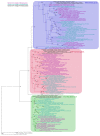
Figures

Similar articles
-
The major clades of MADS-box genes and their role in the development and evolution of flowering plants.Mol Phylogenet Evol. 2003 Dec;29(3):464-89. doi: 10.1016/s1055-7903(03)00207-0. Mol Phylogenet Evol. 2003. PMID: 14615187 Review.
-
Dynamic evolution of FLC/MAF-like genes accompanying Brassicaceae radiation.Ann Bot. 2025 Jun 10:mcaf114. doi: 10.1093/aob/mcaf114. Online ahead of print. Ann Bot. 2025. PMID: 40492332
-
Phylogenomic Synteny Network Analysis of MADS-Box Transcription Factor Genes Reveals Lineage-Specific Transpositions, Ancient Tandem Duplications, and Deep Positional Conservation.Plant Cell. 2017 Jun;29(6):1278-1292. doi: 10.1105/tpc.17.00312. Epub 2017 Jun 5. Plant Cell. 2017. PMID: 28584165 Free PMC article.
-
Phylogenomics reveals surprising sets of essential and dispensable clades of MIKC(c)-group MADS-box genes in flowering plants.J Exp Zool B Mol Dev Evol. 2015 Jun;324(4):353-62. doi: 10.1002/jez.b.22598. Epub 2015 Feb 11. J Exp Zool B Mol Dev Evol. 2015. PMID: 25678468
-
MADS-box genes are involved in floral development and evolution.Acta Biochim Pol. 2001;48(2):351-8. Acta Biochim Pol. 2001. PMID: 11732606 Review.
Cited by
-
Reproductive development in Trithuria submersa (Hydatellaceae: Nymphaeales): the involvement of AGAMOUS-like genes.Planta. 2024 Sep 26;260(5):106. doi: 10.1007/s00425-024-04537-5. Planta. 2024. PMID: 39327272 Free PMC article.
References
-
- Münster T., Pahnke J., Di Rosa A., Kim J.T., Martin W., Saedler H., Theißen G. Floral homeotic genes were recruited from homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants. Proc. Natl. Acad. Sci. USA. 1997;94:2415–2420. doi: 10.1073/pnas.94.6.2415. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

