IRv2-Net: A Deep Learning Framework for Enhanced Polyp Segmentation Performance Integrating InceptionResNetV2 and UNet Architecture with Test Time Augmentation Techniques
- PMID: 37765780
- PMCID: PMC10534485
- DOI: 10.3390/s23187724
IRv2-Net: A Deep Learning Framework for Enhanced Polyp Segmentation Performance Integrating InceptionResNetV2 and UNet Architecture with Test Time Augmentation Techniques
Abstract
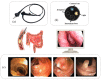
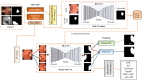
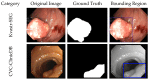
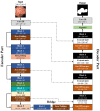
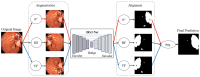
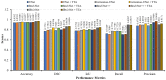
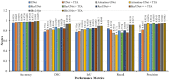
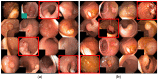
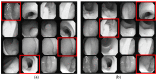
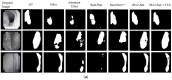
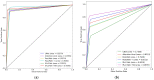
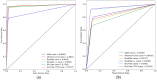
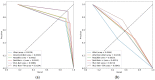
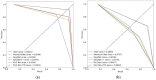
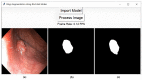
Colorectal polyps in the colon or rectum are precancerous growths that can lead to a more severe disease called colorectal cancer. Accurate segmentation of polyps using medical imaging data is essential for effective diagnosis. However, manual segmentation by endoscopists can be time-consuming, error-prone, and expensive, leading to a high rate of missed anomalies. To solve this problem, an automated diagnostic system based on deep learning algorithms is proposed to find polyps. The proposed IRv2-Net model is developed using the UNet architecture with a pre-trained InceptionResNetV2 encoder to extract most features from the input samples. The Test Time Augmentation (TTA) technique, which utilizes the characteristics of the original, horizontal, and vertical flips, is used to gain precise boundary information and multi-scale image features. The performance of numerous state-of-the-art (SOTA) models is compared using several metrics such as accuracy, Dice Similarity Coefficients (DSC), Intersection Over Union (IoU), precision, and recall. The proposed model is tested on the Kvasir-SEG and CVC-ClinicDB datasets, demonstrating superior performance in handling unseen real-time data. It achieves the highest area coverage in the area under the Receiver Operating Characteristic (ROC-AUC) and area under Precision-Recall (AUC-PR) curves. The model exhibits excellent qualitative testing outcomes across different types of polyps, including more oversized, smaller, over-saturated, sessile, or flat polyps, within the same dataset and across different datasets. Our approach can significantly minimize the number of missed rating difficulties. Lastly, a graphical interface is developed for producing the mask in real-time. The findings of this study have potential applications in clinical colonoscopy procedures and can serve based on further research and development.
Keywords: CVC-ClinicDB; IRv2-Net; Kvasir-SEG; colonoscopy; polyps; segmentation; test time augmentation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Colorectal Cancer: Stages|Cancer.Net. [(accessed on 6 July 2023)]. Available online: https://www.cancer.net/cancer-types/colorectal-cancer/stages.
-
- Hassinger J.P., Hohibar S.D., Pendlirnari R., Dozois E.J., Larson D.W., Cima R.R. Effectiveness of a Multimedia-Based Educational Intervention for Improving Colon Cancer Literacy in Screening Colonoscopy Patients. Dis. Colon Rectum. 2010;53:1301–1307. doi: 10.1007/DCR.0b013e3181e291c0. - DOI - PubMed
-
- Holzheimer R.G., Mannick J.A. Surgical Treatment: Evidence-Based and Problem-Oriented. Zuckschwerdt; Munich, Germany: 2001. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

