EBV and 1q Gains Affect Gene and miRNA Expression in Burkitt Lymphoma
- PMID: 37766215
- PMCID: PMC10537407
- DOI: 10.3390/v15091808
EBV and 1q Gains Affect Gene and miRNA Expression in Burkitt Lymphoma
Abstract
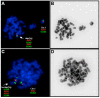
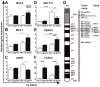
Abnormalities of the long arm of chromosome 1 (1q) represent the most frequent secondary chromosomal aberrations in Burkitt lymphoma (BL) and are observed almost exclusively in EBV-negative BL cell lines (BL-CLs). To verify chromosomal abnormalities, we cytogenetically investigated EBV-negative BL patient material, and to elucidate the 1q gain impact on gene expression, we performed qPCR with six 1q-resident genes and analyzed miRNA expression in BL-CLs. We observed 1q aberrations in the form of duplications, inverted duplications, isodicentric chromosome idic(1)(q10), and the accumulation of 1q12 breakpoints, and we assigned 1q21.2-q32 as a commonly gained region in EBV-negative BL patients. We detected MCL1, ARNT, MLLT11, PDBXIP1, and FCRL5, and 64 miRNAs, showing EBV- and 1q-gain-dependent dysregulation in BL-CLs. We observed MCL1, MLLT11, PDBXIP1, and 1q-resident miRNAs, hsa-miR-9, hsa-miR-9*, hsa-miR-92b, hsa-miR-181a, and hsa-miR-181b, showing copy-number-dependent upregulation in BL-CLs with 1q gains. MLLT11, hsa-miR-181a, hsa-miR-181b, and hsa-miR-183 showed exclusive 1q-gains-dependent and FCRL5, hsa-miR-21, hsa-miR-155, hsa-miR-155*, hsa-miR-221, and hsa-miR-222 showed exclusive EBV-dependent upregulation. We confirmed previous data, e.g., regarding the EBV dependence of hsa-miR-17-92 cluster members, and obtained detailed information considering 1q gains in EBV-negative and EBV-positive BL-CLs. Altogether, our data provide evidence for a non-random involvement of 1q gains in BL and contribute to enlightening and understanding the EBV-negative and EBV-positive BL pathogenesis.
Keywords: 1q gains; Burkitt lymphoma; EBV; FISH; miRNA; qPCR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Johansson B., Brondum-Nielsen K., Billstrom R., Schiodt I., Mitelman F. Translocations between the long arms of chromosomes 1 and 5 in hematologic malignancies are strongly associated with neoplasms of the myeloid lineages. Cancer Genet. Cytogenet. 1997;99:97–101. doi: 10.1016/S0165-4608(97)00198-2. - DOI - PubMed
-
- Itoyama T., Nanjungud G., Chen W., Dyomin V.G., Teruya-Feldstein J., Jhanwar S.C., Zelenetz A.D., Chaganti R.S. Molecular cytogenetic analysis of genomic instability at the 1q12-22 chromosomal site in B-cell non-Hodgkin lymphoma. Genes Chromosomes Cancer. 2002;35:318–328. doi: 10.1002/gcc.10120. - DOI - PubMed
-
- Gunnarsson R., Dilorenzo S., Lundin-Strom K.B., Olsson L., Biloglav A., Lilljebjorn H., Rissler M., Wahlberg P., Lundmark A., Castor A., et al. Mutation, methylation, and gene expression profiles in dup(1q)-positive pediatric B-cell precursor acute lymphoblastic leukemia. Leukemia. 2018;32:2117–2125. doi: 10.1038/s41375-018-0092-2. - DOI - PMC - PubMed
-
- Offit K., Jhanwar S.C., Ladanyi M., Filippa D.A., Chaganti R.S. Cytogenetic analysis of 434 consecutively ascertained specimens of non-Hodgkin’s lymphoma: Correlations between recurrent aberrations, histology, and exposure to cytotoxic treatment. Genes Chromosomes Cancer. 1991;3:189–201. doi: 10.1002/gcc.2870030304. - DOI - PubMed
LinkOut - more resources
Full Text Sources

