Influenza A Virus Infection Alters Lipid Packing and Surface Electrostatic Potential of the Host Plasma Membrane
- PMID: 37766238
- PMCID: PMC10537794
- DOI: 10.3390/v15091830
Influenza A Virus Infection Alters Lipid Packing and Surface Electrostatic Potential of the Host Plasma Membrane
Abstract
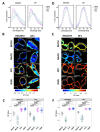
The pathogenesis of influenza A viruses (IAVs) is influenced by several factors, including IAV strain origin and reassortment, tissue tropism and host type. While such factors were mostly investigated in the context of virus entry, fusion and replication, little is known about the viral-induced changes to the host lipid membranes which might be relevant in the context of virion assembly. In this work, we applied several biophysical fluorescence microscope techniques (i.e., Förster energy resonance transfer, generalized polarization imaging and scanning fluorescence correlation spectroscopy) to quantify the effect of infection by two IAV strains of different origin on the plasma membrane (PM) of avian and human cell lines. We found that IAV infection affects the membrane charge of the inner leaflet of the PM. Moreover, we showed that IAV infection impacts lipid-lipid interactions by decreasing membrane fluidity and increasing lipid packing. Because of such alterations, diffusive dynamics of membrane-associated proteins are hindered. Taken together, our results indicate that the infection of avian and human cell lines with IAV strains of different origins had similar effects on the biophysical properties of the PM.
Keywords: biosensors; fluorescence correlation spectroscopy; fluorescence microscopy; fluorescence resonance energy transfer; influenza A virus; lipid packing; membrane fluidity; plasma membrane; quantitative microscopy; spectral imaging.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Cui L., Zheng D., Lee Y.H., Chan T.K., Kumar Y., Ho W.E., Chen J.Z., Tannenbaum S.R., Ong C.N. Metabolomics Investigation Reveals Metabolite Mediators Associated with Acute Lung Injury and Repair in a Murine Model of Influenza Pneumonia. Sci. Rep. 2016;6:26076. doi: 10.1038/srep26076. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

