Genotype and Phenotype Characterization of Rhinolophus sp. Sarbecoviruses from Vietnam: Implications for Coronavirus Emergence
- PMID: 37766303
- PMCID: PMC10536463
- DOI: 10.3390/v15091897
Genotype and Phenotype Characterization of Rhinolophus sp. Sarbecoviruses from Vietnam: Implications for Coronavirus Emergence
Abstract
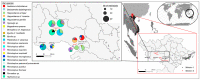
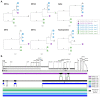
Bats are a major reservoir of zoonotic viruses, including coronaviruses. Since the emergence of SARS-CoV in 2002/2003 in Asia, important efforts have been made to describe the diversity of Coronaviridae circulating in bats worldwide, leading to the discovery of the precursors of epidemic and pandemic sarbecoviruses in horseshoe bats. We investigated the viral communities infecting horseshoe bats living in Northern Vietnam, and report here the first identification of sarbecoviruses in Rhinolophus thomasi and Rhinolophus siamensis bats. Phylogenetic characterization of seven strains of Vietnamese sarbecoviruses identified at least three clusters of viruses. Recombination and cross-species transmission between bats seemed to constitute major drivers of virus evolution. Vietnamese sarbecoviruses were mainly enteric, therefore constituting a risk of spillover for guano collectors or people visiting caves. To evaluate the zoonotic potential of these viruses, we analyzed in silico and in vitro the ability of their RBDs to bind to mammalian ACE2s and concluded that these viruses are likely restricted to their bat hosts. The workflow applied here to characterize the spillover potential of novel sarbecoviruses is of major interest for each time a new virus is discovered, in order to concentrate surveillance efforts on high-risk interfaces.
Keywords: Sarbecovirus; horseshoe bat; spillover; virus evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Anthony S.J., Gilardi K., Menachery V.D., Goldstein T., Ssebide B., Mbabazi R., Navarrete-Macias I., Liang E., Wells H., Hicks A., et al. Further Evidence for Bats as the Evolutionary Source of Middle East Respiratory Syndrome Coronavirus. mBio. 2017;8:e00373-17. doi: 10.1128/mBio.00373-17. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

