Treatment Emergent Dolutegravir Resistance Mutations in Individuals Naïve to HIV-1 Integrase Inhibitors: A Rapid Scoping Review
- PMID: 37766338
- PMCID: PMC10536831
- DOI: 10.3390/v15091932
Treatment Emergent Dolutegravir Resistance Mutations in Individuals Naïve to HIV-1 Integrase Inhibitors: A Rapid Scoping Review
Abstract
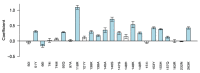
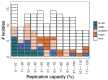
Background: Dolutegravir (DTG)-based antiretroviral therapy (ART) rarely leads to virological failure (VF) and drug resistance in integrase strand transfer inhibitor (INSTI)-naïve persons living with HIV (PLWH). As a result, limited data are available on INSTI-associated drug resistance mutations (DRMs) selected by DTG-containing ART regimens. Methods: We reviewed studies published through July 2023 to identify those reporting emergent major INSTI-associated DRMs in INSTI-naïve PLWH receiving DTG and those containing in vitro DTG susceptibility results using a standardized assay. Results: We identified 36 publications reporting 99 PLWH in whom major nonpolymorphic INSTI-associated DRMs developed on a DTG-containing regimen and 21 publications containing 269 in vitro DTG susceptibility results. DTG-selected DRMs clustered into four largely non-overlapping mutational pathways characterized by mutations at four signature positions: R263K, G118R, N155H, and Q148H/R/K. Eighty-two (82.8%) viruses contained just one signature DRM, including R263K (n = 40), G118R (n = 24), N155H (n = 9), and Q148H/R/K (n = 9). Nine (9.1%) contained ≥1 signature DRM, and eight (8.1%) contained just other DRMs. R263K and G118R were negatively associated with one another and with N155H and Q148H/K/R. R263K alone conferred a median 2.0-fold (IQR: 1.8-2.2) reduction in DTG susceptibility. G118R alone conferred a median 18.8-fold (IQR:14.2-23.4) reduction in DTG susceptibility. N155H alone conferred a median 1.4-fold (IQR: 1.2-1.6) reduction in DTG susceptibility. Q148H/R/K alone conferred a median 0.8-fold (IQR: 0.7-1.1) reduction in DTG susceptibility. Considerably higher levels of reduced susceptibility often occurred when signature DRMs occurred with additional INSTI-associated DRMs. Conclusions: Among INSTI-naïve PLWH with VF and treatment emergent INSTI-associated DRMs, most developed one of four signature DRMs, most commonly R263K or G118R. G118R was associated with a much greater reduction in DTG susceptibility than R263K.
Keywords: Dolutegravir; HIV-1 integrase; antiviral resistance; mutations.
Conflict of interest statement
R.W.S. has received honoraria for participation in advisory boards from Gilead Sciences and GlaxoSmithKline and speaking honoraria from Gilead Sciences and ViiV Healthcare; R.K.G. has received honoraria for participation on advisory boards from Gilead Sciences and GlaxoSmithKline.
Figures


Similar articles
-
Prevalence of Emergent Dolutegravir Resistance Mutations in People Living with HIV: A Rapid Scoping Review.Viruses. 2024 Mar 4;16(3):399. doi: 10.3390/v16030399. Viruses. 2024. PMID: 38543764 Free PMC article.
-
A systematic review of the genetic mechanisms of dolutegravir resistance.J Antimicrob Chemother. 2019 Nov 1;74(11):3135-3149. doi: 10.1093/jac/dkz256. J Antimicrob Chemother. 2019. PMID: 31280314 Free PMC article.
-
Genotypic correlates of resistance to the HIV-1 strand transfer integrase inhibitor cabotegravir.Antiviral Res. 2022 Dec;208:105427. doi: 10.1016/j.antiviral.2022.105427. Epub 2022 Sep 30. Antiviral Res. 2022. PMID: 36191692 Free PMC article.
-
The G118R plus R263K Combination of Integrase Mutations Associated with Dolutegravir-Based Treatment Failure Reduces HIV-1 Replicative Capacity and Integration.Antimicrob Agents Chemother. 2023 May 17;67(5):e0138622. doi: 10.1128/aac.01386-22. Epub 2023 Apr 18. Antimicrob Agents Chemother. 2023. PMID: 37071019 Free PMC article.
-
Integrase strand transfer inhibitor (INSTI)-resistance mutations for the surveillance of transmitted HIV-1 drug resistance.J Antimicrob Chemother. 2020 Jan 1;75(1):170-182. doi: 10.1093/jac/dkz417. J Antimicrob Chemother. 2020. PMID: 31617907 Free PMC article.
Cited by
-
HIV transmission dynamics and population-wide drug resistance in rural South Africa.Nat Commun. 2024 Apr 29;15(1):3644. doi: 10.1038/s41467-024-47254-z. Nat Commun. 2024. PMID: 38684655 Free PMC article.
-
HIV drug resistance to integrase inhibitors in low- and middle-income countries.Nat Med. 2024 Mar;30(3):618-619. doi: 10.1038/s41591-023-02763-0. Nat Med. 2024. PMID: 38263265 Free PMC article. No abstract available.
-
Virological Findings and Treatment Outcomes of Cases That Developed Dolutegravir Resistance in Malawi's National HIV Treatment Program.Viruses. 2023 Dec 23;16(1):29. doi: 10.3390/v16010029. Viruses. 2023. PMID: 38257730 Free PMC article. Review.
-
GPT-4 performance on querying scientific publications: reproducibility, accuracy, and impact of an instruction sheet.BMC Med Res Methodol. 2024 Jun 25;24(1):139. doi: 10.1186/s12874-024-02253-y. BMC Med Res Methodol. 2024. PMID: 38918736 Free PMC article.
-
Patterns of HIV-1 Drug Resistance Observed Through Geospatial Analysis of Routine Diagnostic Testing in KwaZulu-Natal, South Africa.Viruses. 2024 Oct 19;16(10):1634. doi: 10.3390/v16101634. Viruses. 2024. PMID: 39459966 Free PMC article.
References
-
- Gandhi R.T., Bedimo R., Hoy J.F., Landovitz R.J., Smith D.M., Eaton E.F., Lehmann C., Springer S.A., Sax P.E., Thompson M.A., et al. Antiretroviral Drugs for Treatment and Prevention of HIV Infection in Adults: 2022 Recommendations of the International Antiviral Society–USA Panel. JAMA. 2022;329:63. doi: 10.1001/jama.2022.22246. - DOI - PubMed
-
- World Health Organization Consolidated Guidelines on HIV Prevention, Testing, Treatment, Service Delivery and Monitoring: Recommendations for a Public Health Approach. [(accessed on 17 August 2022)]. Available online: https://www.who.int/publications-detail-redirect/9789240031593. - PubMed
-
- Tzou P.L., Rhee S.-Y., Descamps D., Clutter D.S., Hare B., Mor O., Grude M., Parkin N., Jordan M.R., Bertagnolio S., et al. Integrase Strand Transfer Inhibitor (INSTI)-Resistance Mutations for the Surveillance of Transmitted HIV-1 Drug Resistance. J. Antimicrob. Chemother. 2020;75:170–182. doi: 10.1093/jac/dkz417. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

