Real-Time Analysis of SARS-CoV-2-Induced Cytolysis Reveals Distinct Variant-Specific Replication Profiles
- PMID: 37766343
- PMCID: PMC10537736
- DOI: 10.3390/v15091937
Real-Time Analysis of SARS-CoV-2-Induced Cytolysis Reveals Distinct Variant-Specific Replication Profiles
Abstract
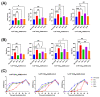
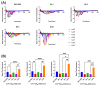
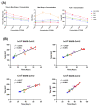
The ability of each new SARS-CoV-2 variant to evade host humoral immunity is the focus of intense research. Each variant may also harbor unique replication capabilities relevant for disease and transmission. Here, we demonstrate a new approach to assessing viral replication kinetics using real-time cell analysis (RTCA). Virus-induced cell death is measured in real time as changes in electrical impedance through cell monolayers while images are acquired at defined intervals via an onboard microscope and camera. Using this system, we quantified replication kinetics of five clinically important viral variants: WA1/2020 (ancestral), Delta, and Omicron subvariants BA.1, BA.4, and BA.5. Multiple measures proved useful in variant replication comparisons, including the elapsed time to, and the slope at, the maximum rate of cell death. Important findings include significantly weaker replication kinetics of BA.1 by all measures, while BA.5 harbored replication kinetics at or near ancestral levels, suggesting evolution to regain replicative capacity, and both an altered profile of cell killing and enhanced fusogenicity of the Delta variant. Together, these data show that RTCA is a robust method to assess replicative capacity of any given SARS-CoV-2 variant rapidly and quantitatively, which may be useful in assessment of newly emerging variants.
Keywords: SARS-CoV-2 variants; real-time cell analysis; viral replication.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole A., Southgate J., Johnson R., Jackson B., Nascimento F.F., et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. - DOI - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

