The Disruption of a Nuclear Export Signal in the C-Terminus of the Herpes Simplex Virus 1 Determinant of Pathogenicity UL24 Protein Leads to a Syncytial Plaque Phenotype
- PMID: 37766377
- PMCID: PMC10535440
- DOI: 10.3390/v15091971
The Disruption of a Nuclear Export Signal in the C-Terminus of the Herpes Simplex Virus 1 Determinant of Pathogenicity UL24 Protein Leads to a Syncytial Plaque Phenotype
Abstract
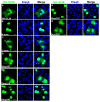
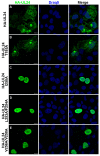
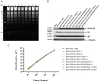
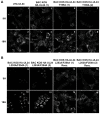
UL24 of herpes simplex virus 1 (HSV-1) has been shown to be a determinant of pathogenesis in mouse models of infection. The N-terminus of UL24 localizes to the nucleus and drives the redistribution of nucleolin and B23. In contrast, when expressed alone, the C-terminal domain of UL24 accumulates in the Golgi apparatus; its importance during infection is unknown. We generated a series of mammalian expression vectors encoding UL24 with nested deletions in the C-terminal domain. Interestingly, enhanced nuclear staining was observed for several UL24-deleted forms in transient transfection assays. The substitution of a threonine phosphorylation site had no effect on UL24 localization or viral titers in cell culture. In contrast, mutations targeting a predicted nuclear export signal (NES) significantly enhanced nuclear localization, indicating that UL24 is able to shuttle between the nucleus and the cytoplasm. Recombinant viruses that encode UL24-harboring substitutions in the NES led to the accumulation of UL24 in the nucleus. Treatment with the CRM-1-specific inhibitor leptomycin B blocked the nuclear export of UL24 in transfected cells but not in the context of infection. Viruses encoding UL24 with NES mutations resulted in a syncytial phenotype, but viral yield was unaffected. These results are consistent with a role for HSV-1 UL24 in late cytoplasmic events in HSV-1 replication.
Keywords: UL24; herpes simplex virus; nuclear export signal; syncytia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
A Mutation in the UL24 Gene Abolishes Expression of the Newly Identified UL24.5 Protein of Herpes Simplex Virus 1 and Leads to an Increase in Pathogenicity in Mice.J Virol. 2018 Sep 26;92(20):e00671-18. doi: 10.1128/JVI.00671-18. Print 2018 Oct 15. J Virol. 2018. PMID: 30068651 Free PMC article.
-
The conserved N-terminal domain of herpes simplex virus 1 UL24 protein is sufficient to induce the spatial redistribution of nucleolin.J Gen Virol. 2008 May;89(Pt 5):1142-1151. doi: 10.1099/vir.0.83573-0. J Gen Virol. 2008. PMID: 18420791
-
Involvement of the UL24 protein in herpes simplex virus 1-induced dispersal of B23 and in nuclear egress.Virology. 2011 Apr 10;412(2):341-8. doi: 10.1016/j.virol.2011.01.016. Epub 2011 Feb 12. Virology. 2011. PMID: 21316727
-
Mechanism of herpesvirus UL24 protein regulating viral immune escape and virulence.Front Microbiol. 2023 Sep 22;14:1268429. doi: 10.3389/fmicb.2023.1268429. eCollection 2023. Front Microbiol. 2023. PMID: 37808279 Free PMC article. Review.
-
UL24 herpesvirus determinants of pathogenesis: Roles in virus-host interactions.Virology. 2025 Feb;603:110376. doi: 10.1016/j.virol.2024.110376. Epub 2024 Dec 24. Virology. 2025. PMID: 39765022 Review.
Cited by
-
Comparative Review of the Conserved UL24 Protein Family in Herpesviruses.Int J Mol Sci. 2024 Oct 19;25(20):11268. doi: 10.3390/ijms252011268. Int J Mol Sci. 2024. PMID: 39457049 Free PMC article. Review.
-
Herpes Simplex Virus Type 2 Blocks IFN-β Production through the Viral UL24 N-Terminal Domain-Mediated Inhibition of IRF-3 Phosphorylation.Viruses. 2024 Oct 11;16(10):1601. doi: 10.3390/v16101601. Viruses. 2024. PMID: 39459934 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

