Riluzole Suppresses Growth and Enhances Response to Endocrine Therapy in ER+ Breast Cancer
- PMID: 37766843
- PMCID: PMC10521904
- DOI: 10.1210/jendso/bvad117
Riluzole Suppresses Growth and Enhances Response to Endocrine Therapy in ER+ Breast Cancer
Abstract
Background: Resistance to endocrine therapy in estrogen receptor-positive (ER+) breast cancer remains a significant clinical problem. Riluzole is FDA-approved for the treatment of amyotrophic lateral sclerosis. A benzothiazole-based glutamate release inhibitor with several context-dependent mechanism(s) of action, riluzole has shown antitumor activity in multiple malignancies, including melanoma, glioblastoma, and breast cancer. We previously reported that the acquisition of tamoxifen resistance in a cellular model of invasive lobular breast cancer is accompanied by the upregulation of GRM mRNA expression and growth inhibition by riluzole.
Methods: We tested the ability of riluzole to reduce cell growth, alone and in combination with endocrine therapy, in a diverse set of ER+ invasive ductal and lobular breast cancer-derived cell lines, primary breast tumor explant cultures, and the estrogen-independent, ESR1-mutated invasive lobular breast cancer patient-derived xenograft model HCI-013EI.
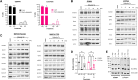
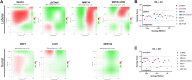
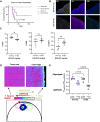
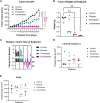
Results: Single-agent riluzole suppressed the growth of ER+ invasive ductal and lobular breast cancer cell lines in vitro, inducing a histologic subtype-associated cell cycle arrest (G0-G1 for ductal, G2-M for lobular). Riluzole induced apoptosis and ferroptosis and reduced phosphorylation of multiple prosurvival signaling molecules, including Akt/mTOR, CREB, and Fak/Src family kinases. Riluzole, in combination with either fulvestrant or 4-hydroxytamoxifen, additively suppressed ER+ breast cancer cell growth in vitro. Single-agent riluzole significantly inhibited HCI-013EI patient-derived xenograft growth in vivo, and the combination of riluzole plus fulvestrant significantly reduced proliferation in ex vivo primary breast tumor explant cultures.
Conclusion: Riluzole may offer therapeutic benefits in diverse ER+ breast cancers, including lobular breast cancer.
Keywords: estrogen receptor; fulvestrant; invasive lobular breast cancer; riluzole.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Endocrine Society.
Figures




Comment in
-
A Path to Precision Metabolic Treatment in Breast Cancer: Riluzole, Glutamate Signaling, and Invasive Lobular Carcinoma.J Endocr Soc. 2023 Dec 26;8(2):bvad171. doi: 10.1210/jendso/bvad171. eCollection 2024 Jan 5. J Endocr Soc. 2023. PMID: 38193025 Free PMC article. No abstract available.
References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17‐48. - PubMed
-
- Burstein HJ, Lacchetti C, Anderson H, et al. Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: ASCO clinical practice guideline focused update. J Clin Oncol. 2019;37(5):423‐438. - PubMed
-
- Desmedt C, Zoppoli G, Gundem G, et al. Genomic characterization of primary invasive lobular breast cancer. J Clin Oncol. 2016;34(16):1872‐1881. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
