Global quantitative understanding of non-equilibrium cell fate decision-making in response to pheromone
- PMID: 37766979
- PMCID: PMC10520453
- DOI: 10.1016/j.isci.2023.107885
Global quantitative understanding of non-equilibrium cell fate decision-making in response to pheromone
Abstract
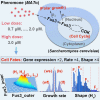
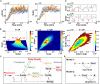
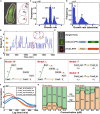
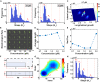
Cell-cycle arrest and polarized growth are commonly used to characterize the response of yeast to pheromone. However, the quantitative decision-making processes underlying time-dependent changes in cell fate remain unclear. In this study, we conducted single-cell level experiments to observe multidimensional responses, uncovering diverse fates of yeast cells. Multiple states are revealed, along with the kinetic switching rates and pathways among them, giving rise to a quantitative landscape of mating response. To quantify the experimentally observed cell fates, we developed a theoretical framework based on non-equilibrium landscape and flux theory. Additionally, we performed stochastic simulations of biochemical reactions to elucidate signal transduction and cell growth. Notably, our experimental findings have provided the first global quantitative evidence of the real-time synchronization between intracellular signaling, physiological growth, and morphological functions. These results validate the proposed underlying mechanism governing the emergence of multiple cell fate states. This study introduces an emerging mechanistic approach to understand non-equilibrium cell fate decision-making in response to pheromone.
Keywords: Biological sciences; Cell biology; Cellular physiology.
© 2023 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Quantifying nonequilibrium dynamics and thermodynamics of cell fate decision making in yeast under pheromone induction.Biophys Rev (Melville). 2023 Sep 13;4(3):031401. doi: 10.1063/5.0157759. eCollection 2023 Sep. Biophys Rev (Melville). 2023. PMID: 38510708 Free PMC article.
-
Landscape and flux quantify the stochastic transition dynamics for p53 cell fate decision.J Chem Phys. 2021 Jan 14;154(2):025101. doi: 10.1063/5.0030558. J Chem Phys. 2021. PMID: 33445890
-
Reconstructing the regulatory circuit of cell fate determination in yeast mating response.PLoS Comput Biol. 2017 Jul 24;13(7):e1005671. doi: 10.1371/journal.pcbi.1005671. eCollection 2017 Jul. PLoS Comput Biol. 2017. PMID: 28742153 Free PMC article.
-
Processes on the emergent landscapes of biochemical reaction networks and heterogeneous cell population dynamics: differentiation in living matters.J R Soc Interface. 2017 May;14(130):20170097. doi: 10.1098/rsif.2017.0097. J R Soc Interface. 2017. PMID: 28490602 Free PMC article. Review.
-
Single-cell approaches to cell competition: High-throughput imaging, machine learning and simulations.Semin Cancer Biol. 2020 Jun;63:60-68. doi: 10.1016/j.semcancer.2019.05.007. Epub 2019 May 18. Semin Cancer Biol. 2020. PMID: 31108201 Review.
Cited by
-
Quantifying nonequilibrium dynamics and thermodynamics of cell fate decision making in yeast under pheromone induction.Biophys Rev (Melville). 2023 Sep 13;4(3):031401. doi: 10.1063/5.0157759. eCollection 2023 Sep. Biophys Rev (Melville). 2023. PMID: 38510708 Free PMC article.
-
Quantifying Landscape-Flux via Single-Cell Transcriptomics Uncovers the Underlying Mechanism of Cell Cycle.Adv Sci (Weinh). 2024 Apr;11(16):e2308879. doi: 10.1002/advs.202308879. Epub 2024 Feb 14. Adv Sci (Weinh). 2024. PMID: 38353329 Free PMC article.
References
-
- Berg H.C., Brown D.A. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–504. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

