Bst polymerase - a humble relative of Taq polymerase
- PMID: 37767105
- PMCID: PMC10520511
- DOI: 10.1016/j.csbj.2023.09.008
Bst polymerase - a humble relative of Taq polymerase
Abstract
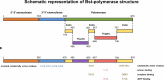
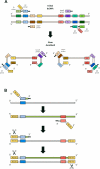
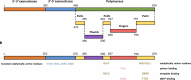
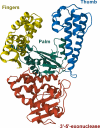
DNA polymerases are a superfamily of enzymes synthesizing DNA using DNA as a template. They are essential for nucleic acid metabolism and for DNA replication and repair. Modern biotechnology and molecular diagnostics rely heavily on DNA polymerases in analyzing nucleic acids. Among a variety of discovered DNA polymerases, Bst polymerase, a large fragment of DNA polymerase I from Geobacillus stearothermophilus, is one of the most commonly used but is not as well studied as Taq polymerase. The ability of Bst polymerase to displace an upstream DNA strand during synthesis, coupled with its moderate thermal stability, has provided the basis for several isothermal DNA amplification methods, including LAMP, WGA, RCA, and many others. Bst polymerase is one of the key components defining the robustness and analytical characteristics of diagnostic test systems based on isothermal amplification. Here, we present an overview of the biochemical and structural features of Bst polymerase and provide information on its mutated analogs.
Keywords: Bst polymerase; DNA polymerase; Directed evolution; Fidelity; Fusion proteins; Processivity; Site-directed mutagenesis; Strand displacement.
© 2023 The Authors.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
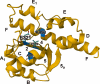
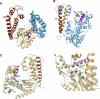
Similar articles
-
Derivatives of Bst-like Gss-polymerase with improved processivity and inhibitor tolerance.Nucleic Acids Res. 2017 Sep 19;45(16):9595-9610. doi: 10.1093/nar/gkx645. Nucleic Acids Res. 2017. PMID: 28934494 Free PMC article.
-
Characterization and engineering of a DNA polymerase reveals a single amino-acid substitution in the fingers subdomain to increase strand-displacement activity of A-family prokaryotic DNA polymerases.BMC Mol Cell Biol. 2019 Aug 9;20(1):31. doi: 10.1186/s12860-019-0216-1. BMC Mol Cell Biol. 2019. PMID: 31399021 Free PMC article.
-
The Influence of Reaction Conditions on DNA Multimerization During Isothermal Amplification with Bst exo- DNA Polymerase.Appl Biochem Biotechnol. 2020 Feb;190(2):758-771. doi: 10.1007/s12010-019-03127-6. Epub 2019 Sep 7. Appl Biochem Biotechnol. 2020. PMID: 31493160
-
DNA Polymerases for Whole Genome Amplification: Considerations and Future Directions.Int J Mol Sci. 2023 May 26;24(11):9331. doi: 10.3390/ijms24119331. Int J Mol Sci. 2023. PMID: 37298280 Free PMC article. Review.
-
Loop-mediated isothermal amplification of DNA (LAMP): a new diagnostic tool lights the world of diagnosis of animal and human pathogens: a review.Pak J Biol Sci. 2014 Jan 15;17(2):151-66. doi: 10.3923/pjbs.2014.151.166. Pak J Biol Sci. 2014. PMID: 24783797 Review.
Cited by
-
Development of reverse transcription loop-mediated isothermal amplification-based assay for rapid and specific detection of human fungal pathogen, Candida auris.Curr Med Mycol. 2024 Nov 14;10:e2024.345284.1572. doi: 10.22034/cmm.2024.345284.1572. eCollection 2024. Curr Med Mycol. 2024. PMID: 40584605 Free PMC article.
-
Point-of-Care Diagnostic Test for Rapid Detection of Infectious Laryngotracheitis Virus by Loop-Mediated Isothermal Amplification and Nanoprobes.Int J Mol Sci. 2025 Feb 25;26(5):1971. doi: 10.3390/ijms26051971. Int J Mol Sci. 2025. PMID: 40076597 Free PMC article.
-
Design and Optimization of Isothermal Gene Amplification for Generation of High-Gain Oligonucleotide Products by MicroRNAs.ACS Meas Sci Au. 2024 Nov 8;4(6):737-750. doi: 10.1021/acsmeasuresciau.4c00063. eCollection 2024 Dec 18. ACS Meas Sci Au. 2024. PMID: 39713023 Free PMC article.
-
Efficient Genome Editing with Chimeric Oligonucleotide-Directed Editing.bioRxiv [Preprint]. 2024 Jul 10:2024.07.09.602710. doi: 10.1101/2024.07.09.602710. bioRxiv. 2024. PMID: 39026836 Free PMC article. Preprint.
-
Loop-Mediated Isothermal Amplification Assay for Visual Detection of Salmonella enterica Serovar Typhimurium in Food Animal Meat Products.Foods. 2025 May 13;14(10):1731. doi: 10.3390/foods14101731. Foods. 2025. PMID: 40428511 Free PMC article.
References
-
- Brian K. Maples, Rebecca C. Holmberg, Andrew P. Miller, Jarrod Provins, Richard B. Roth JM. Nicking and extension amplification reaction for the exponential amplification of nucleic acids. US20090081670A1, 2008.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
