Mild dehydration effects on the murine kidney single-nucleus transcriptome and chromatin accessibility
- PMID: 37767569
- PMCID: PMC11550884
- DOI: 10.1152/ajprenal.00161.2023
Mild dehydration effects on the murine kidney single-nucleus transcriptome and chromatin accessibility
Erratum in
-
Corrigendum for Huynh et al., volume 325, 2023, p. F730.Am J Physiol Renal Physiol. 2024 Apr 1;326(4):F642. doi: 10.1152/ajprenal.00161.2023_COR. Am J Physiol Renal Physiol. 2024. PMID: 38569020 Free PMC article. No abstract available.
Abstract
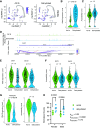
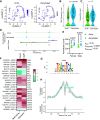
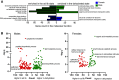
Daily, we may experience mild dehydration with a rise in plasma osmolality that triggers the release of vasopressin. Although the effect of dehydration is well characterized in collecting duct principal cells (CDPCs), we hypothesized that mild dehydration (<12 h) results in many kidney cell-specific changes in transcriptomes and chromatin accessibility. Single-nucleus (sn) multiome (RNA-assay for transposase-accessible chromatin) sequencing and bulk RNA sequencing of kidneys from male and female mice that were mildly water deprived or not were compared. Water-deprived mice had a significant increase in plasma osmolality. sn-multiome-seq resulted in 19,837 nuclei that were annotated into 33 clusters. In CDPCs, aquaporin 2 (Aqp2) and aquaporin 3 (Apq3) were greater in dehydrated mice, but there were novel genes like gremlin 2 (Grem2; a cytokine) that were increased compared with ad libitum mice. The transcription factor cAMP-responsive element modulator (Crem) was greater in CDPCs of dehydrated mice, and the Crem DNA motif was more accessible. There were hundreds of sex- and dehydration-specific differentially expressed genes (DEGs) throughout the kidney, especially in the proximal tubules and thin limbs. In male mice, DEGs were enriched in pathways related to lipid metabolism, whereas female DEGs were enriched in organic acid metabolism. Many highly expressed genes had a positive correlation with increased chromatin accessibility, and mild dehydration exerted many transcriptional changes that we detected at the chromatin level. Even with a rise in plasma osmolality, male and female kidneys have distinct transcriptomes suggesting that there may be diverse mechanisms used to remain in fluid balance.NEW & NOTEWORTHY The kidney consists of >30 cell types that work collectively to maintain fluid-electrolyte balance. Kidney single-nucleus transcriptomes and chromatin accessibility profiles from male and female control (ad libitum water and food) or mildly dehydrated mice (ad libitum food, water deprivation) were determined. Mild dehydration caused hundreds of cell- and sex-specific transcriptomic changes, even though the kidney function to conserve water was the same.
Keywords: chromatin; dehydration; kidney; sex differences; transcriptome.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures





References
-
- Chen L, Jung HJ, Datta A, Park E, Poll BG, Kikuchi H, Leo KT, Mehta Y, Lewis S, Khundmiri SJ, Khan S, Chou CL, Raghuram V, Yang CR, Knepper MA. Systems biology of the vasopressin V2 receptor: new tools for discovery of molecular actions of a GPCR. Annu Rev Pharmacol Toxicol 62: 595–616, 2022. doi: 10.1146/annurev-pharmtox-052120-011012. - DOI - PMC - PubMed
-
- Dumas SJ, Meta E, Borri M, Goveia J, Rohlenova K, Conchinha NV, Falkenberg K, Teuwen LA, de Rooij L, Kalucka J, Chen R, Khan S, Taverna F, Lu W, Parys M, De Legher C, Vinckier S, Karakach TK, Schoonjans L, Lin L, Bolund L, Dewerchin M, Eelen G, Rabelink TJ, Li X, Luo Y, Carmeliet P. Single-cell RNA sequencing reveals renal endothelium heterogeneity and metabolic adaptation to water deprivation. J Am Soc Nephrol 31: 118–138, 2020. doi: 10.1681/ASN.2019080832. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

