An unusual case of thalassemia intermedia with inheritable complex repeats detected by single-molecule optical mapping
- PMID: 37767576
- PMCID: PMC10905065
- DOI: 10.3324/haematol.2023.282902
An unusual case of thalassemia intermedia with inheritable complex repeats detected by single-molecule optical mapping
Figures

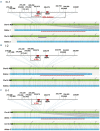
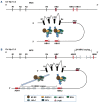
References
-
- Chiba K, Shiraishi Y, Nagata Y, et al. Genomon ITDetector: a tool for somatic internal tandem duplication detection from cancer genome sequencing data. Bioinformatics. 2015;31(1):116-118. - PubMed
-
- Shang X, Xu XM. Update in the genetics of thalassemia: what clinicians need to know. Best Pract Res Clin Obstet Gynaecol. 2017;39:3-15. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

