Chemokine-like Orion is involved in the transformation of glial cells into phagocytes in different developmental neuronal remodeling paradigms
- PMID: 37767633
- PMCID: PMC10565233
- DOI: 10.1242/dev.201633
Chemokine-like Orion is involved in the transformation of glial cells into phagocytes in different developmental neuronal remodeling paradigms
Abstract
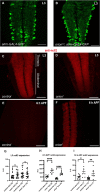
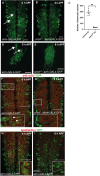
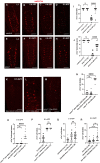
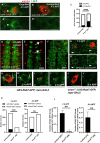
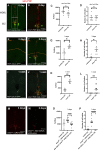
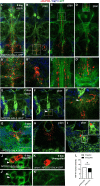
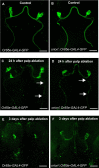
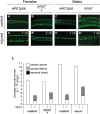
During animal development, neurons often form exuberant or inappropriate axons and dendrites at early stages, followed by the refinement of neuronal circuits at late stages. Neural circuit refinement leads to the production of neuronal debris in the form of neuronal cell corpses, fragmented axons and dendrites, and pruned synapses requiring disposal. Glial cells act as predominant phagocytes during neuronal remodeling and degeneration, and crucial signaling pathways between neurons and glia are necessary for the execution of phagocytosis. Chemokine-like mushroom body neuron-secreted Orion is essential for astrocyte infiltration into the γ axon bundle leading to γ axon pruning. Here, we show a role of Orion in debris engulfment and phagocytosis in Drosophila. Interestingly, Orion is involved in the overall transformation of astrocytes into phagocytes. In addition, analysis of several neuronal paradigms demonstrates the role of Orion in eliminating both peptidergic vCrz+ and PDF-Tri neurons via additional phagocytic glial cells like cortex and/or ensheathing glia. Our results suggest that Orion is essential for phagocytic activation of astrocytes, cortex and ensheathing glia, and point to Orion as a trigger of glial infiltration, engulfment and phagocytosis.
Keywords: Drosophila; Axon and cell body remodeling; Chemokine-like Orion; Glia; Neuron; Phagocytosis.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures
Similar articles
-
Neuron-to-glia and glia-to-glia signaling directs critical period experience-dependent synapse pruning.Front Cell Dev Biol. 2025 Feb 18;13:1540052. doi: 10.3389/fcell.2025.1540052. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40040788 Free PMC article. Review.
-
Astrocytes engage unique molecular programs to engulf pruned neuronal debris from distinct subsets of neurons.Genes Dev. 2014 Jan 1;28(1):20-33. doi: 10.1101/gad.229518.113. Epub 2013 Dec 20. Genes Dev. 2014. PMID: 24361692 Free PMC article.
-
Neuron-glia crosstalk in neuronal remodeling and degeneration: Neuronal signals inducing glial cell phagocytic transformation in Drosophila.Bioessays. 2022 May;44(5):e2100254. doi: 10.1002/bies.202100254. Epub 2022 Mar 22. Bioessays. 2022. PMID: 35315125
-
Axonal chemokine-like Orion induces astrocyte infiltration and engulfment during mushroom body neuronal remodeling.Nat Commun. 2021 Mar 23;12(1):1849. doi: 10.1038/s41467-021-22054-x. Nat Commun. 2021. PMID: 33758182 Free PMC article.
-
Nuclear receptors and Drosophila neuronal remodeling.Biochim Biophys Acta. 2015 Feb;1849(2):187-95. doi: 10.1016/j.bbagrm.2014.05.024. Epub 2014 Jun 2. Biochim Biophys Acta. 2015. PMID: 24882358 Review.
Cited by
-
Astrocyte-dependent local neurite pruning in Beat-Va neurons.J Cell Biol. 2025 Jan 6;224(1):e202312043. doi: 10.1083/jcb.202312043. Epub 2024 Dec 9. J Cell Biol. 2025. PMID: 39652106 Free PMC article.
-
Neuron-to-glia and glia-to-glia signaling directs critical period experience-dependent synapse pruning.Front Cell Dev Biol. 2025 Feb 18;13:1540052. doi: 10.3389/fcell.2025.1540052. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40040788 Free PMC article. Review.
-
Neuron-to-glia signaling drives critical period experience-dependent synapse pruning.Sci Rep. 2025 Jul 16;15(1):25744. doi: 10.1038/s41598-025-11528-3. Sci Rep. 2025. PMID: 40670566 Free PMC article.
-
Glia multitask to compensate for neighboring glial cell dysfunction.bioRxiv [Preprint]. 2024 Sep 10:2024.09.06.611719. doi: 10.1101/2024.09.06.611719. bioRxiv. 2024. Update in: Glia. 2025 Aug 6. doi: 10.1002/glia.70072. PMID: 39314422 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

