Oncogenic KRAS Drives Lipofibrogenesis to Promote Angiogenesis and Colon Cancer Progression
- PMID: 37768068
- PMCID: PMC10807546
- DOI: 10.1158/2159-8290.CD-22-1467
Oncogenic KRAS Drives Lipofibrogenesis to Promote Angiogenesis and Colon Cancer Progression
Abstract
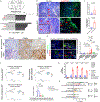
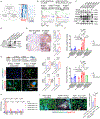
Oncogenic KRAS (KRAS*) contributes to many cancer hallmarks. In colorectal cancer, KRAS* suppresses antitumor immunity to promote tumor invasion and metastasis. Here, we uncovered that KRAS* transforms the phenotype of carcinoma-associated fibroblasts (CAF) into lipid-laden CAFs, promoting angiogenesis and tumor progression. Mechanistically, KRAS* activates the transcription factor CP2 (TFCP2) that upregulates the expression of the proadipogenic factors BMP4 and WNT5B, triggering the transformation of CAFs into lipid-rich CAFs. These lipid-rich CAFs, in turn, produce VEGFA to spur angiogenesis. In KRAS*-driven colorectal cancer mouse models, genetic or pharmacologic neutralization of TFCP2 reduced lipid-rich CAFs, lessened tumor angiogenesis, and improved overall survival. Correspondingly, in human colorectal cancer, lipid-rich CAF and TFCP2 signatures correlate with worse prognosis. This work unveils a new role for KRAS* in transforming CAFs, driving tumor angiogenesis and disease progression, providing an actionable therapeutic intervention for KRAS*-driven colorectal cancer.
Significance: This study identified a molecular mechanism contributing to KRAS*-driven colorectal cancer progression via fibroblast transformation in the tumor microenvironment to produce VEGFA driving tumor angiogenesis. In preclinical models, targeting the KRAS*-TFCP2-VEGFA axis impaired tumor progression, revealing a potential novel therapeutic option for patients with KRAS*-driven colorectal cancer. This article is featured in Selected Articles from This Issue, p. 2489.
©2023 American Association for Cancer Research.
Conflict of interest statement
Figures



References
-
- Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, and Bouvier AM (2006). Epidemiology And Management Of Liver Metastases From Colorectal Cancer. Ann Surg 244, 254–259. 10.1097/01.sla.0000217629.94941.cf. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

