Quantitative detection and reduction of potentially pathogenic bacterial groups of Aeromonas, Arcobacter, Klebsiella pneumoniae species complex, and Mycobacterium in wastewater treatment facilities
- PMID: 37768925
- PMCID: PMC10538766
- DOI: 10.1371/journal.pone.0291742
Quantitative detection and reduction of potentially pathogenic bacterial groups of Aeromonas, Arcobacter, Klebsiella pneumoniae species complex, and Mycobacterium in wastewater treatment facilities
Abstract
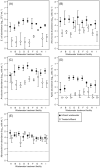
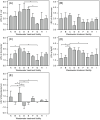
Water quality parameters influence the abundance of pathogenic bacteria. The genera Aeromonas, Arcobacter, Klebsiella, and Mycobacterium are among the representative pathogenic bacteria identified in wastewater. However, information on the correlations between water quality and the abundance of these bacteria, as well as their reduction rate in existing wastewater treatment facilities (WTFs), is lacking. Hence, this study aimed to determine the abundance and reduction rates of these bacterial groups in WTFs. Sixty-eight samples (34 influent and 34 non-disinfected, treated, effluent samples) were collected from nine WTFs in Japan and Thailand. 16S rRNA gene amplicon sequencing analysis revealed the presence of Aeromonas, Arcobacter, and Mycobacterium in all influent wastewater and treated effluent samples. Quantitative real-time polymerase chain reaction (qPCR) was used to quantify the abundance of Aeromonas, Arcobacter, Klebsiella pneumoniae species complex (KpSC), and Mycobacterium. The geometric mean abundances of Aeromonas, Arcobacter, KpSC, and Mycobacterium in the influent wastewater were 1.2 × 104-2.4 × 105, 1.0 × 105-4.5 × 106, 3.6 × 102-4.3 × 104, and 6.9 × 103-5.5 × 104 cells mL-1, respectively, and their average log reduction values were 0.77-2.57, 1.00-3.06, 1.35-3.11, and -0.67-1.57, respectively. Spearman's rank correlation coefficients indicated significant positive or negative correlations between the abundances of the potentially pathogenic bacterial groups and Escherichia coli as well as water quality parameters, namely, chemical/biochemical oxygen demand, total nitrogen, nitrate-nitrogen, nitrite-nitrogen, ammonium-nitrogen, suspended solids, volatile suspended solids, and oxidation-reduction potential. This study provides valuable information on the development and appropriate management of WTFs to produce safe, hygienic water.
Copyright: © 2023 Aoki et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Jones ER, van Vliet MTH, Qadir M, Bierkens MFP. Country-level and gridded estimates of wastewater production, collection, treatment and reuse. Earth Syst. Sci. Data 2021;13(2):237–254. doi: 10.5194/essd-13-237-2021 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

