Precision medicine in the era of multi-omics: can the data tsunami guide rational treatment decision?
- PMID: 37769400
- PMCID: PMC10539962
- DOI: 10.1016/j.esmoop.2023.101642
Precision medicine in the era of multi-omics: can the data tsunami guide rational treatment decision?
Abstract
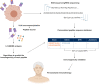
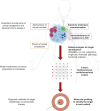
Precision medicine for cancer is rapidly moving to an approach that integrates multiple dimensions of the biology in order to model mechanisms of cancer progression in each patient. The discovery of multiple drivers per tumor challenges medical decision that faces several treatment options. Drug sensitivity depends on the actionability of the target, its clonal or subclonal origin and coexisting genomic alterations. Sequencing has revealed a large diversity of drivers emerging at treatment failure, which are potential targets for clinical trials or drug repurposing. To effectively prioritize therapies, it is essential to rank genomic alterations based on their proven actionability. Moving beyond primary drivers, the future of precision medicine necessitates acknowledging the intricate spatial and temporal heterogeneity inherent in cancer. The advent of abundant complex biological data will make artificial intelligence algorithms indispensable for thorough analysis. Here, we will discuss the advancements brought by the use of high-throughput genomics, the advantages and limitations of precision medicine studies and future perspectives in this field.
Keywords: ESCAT; NGS; OncoKB; high-throughput genomics; precision medicine.
Copyright © 2023 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures


References
-
- Yates L.R., Seoane J., Le Tourneau C., et al. The European Society for Medical Oncology (ESMO) precision medicine glossary. Ann Oncol. 2018;29:30–35. - PubMed
-
- Druker B.J., Talpaz M., Resta D.J., et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

