Ablation of ERO1A induces lethal endoplasmic reticulum stress responses and immunogenic cell death to activate anti-tumor immunity
- PMID: 37769655
- PMCID: PMC10591028
- DOI: 10.1016/j.xcrm.2023.101206
Ablation of ERO1A induces lethal endoplasmic reticulum stress responses and immunogenic cell death to activate anti-tumor immunity
Abstract
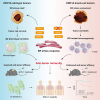
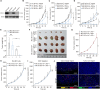
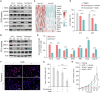
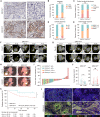
Immunophenotyping of the tumor microenvironment (TME) is essential for enhancing immunotherapy efficacy. However, strategies for characterizing the TME exhibit significant heterogeneity. Here, we show that endoplasmic reticular oxidoreductase-1α (ERO1A) mediates an immune-suppressive TME and attenuates the response to PD-1 blockade. Ablation of ERO1A in tumor cells substantially incites anti-tumor T cell immunity and promotes the efficacy of aPD-1 in therapeutic models. Single-cell RNA-sequencing analyses confirm that ERO1A correlates with immunosuppression and dysfunction of CD8+ T cells along anti-PD-1 treatment. In human lung cancer, high ERO1A expression is associated with a higher risk of recurrence following neoadjuvant immunotherapy. Mechanistically, ERO1A ablation impairs the balance between IRE1α and PERK signaling activities and induces lethal unfolded protein responses in tumor cells undergoing endoplasmic reticulum stress, thereby enhancing anti-tumor immunity via immunogenic cell death. These findings reveal how tumor ERO1A induces immunosuppression, highlighting its potential as a therapeutic target for cancer immunotherapy.
Keywords: ERO1A; endoplasmic reticulum stress response; immune target; immunotherapy; tumor microenvironment.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., Coussens L.M., Gabrilovich D.I., Ostrand-Rosenberg S., Hedrick C.C., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

