Establishment and application of a point-of-care testing and diagnosis method for early immediate expression gene IE1 of cytomegalovirus in maternal urine based on isothermal amplification
- PMID: 37769815
- PMCID: PMC10579523
- DOI: 10.1016/j.virusres.2023.199229
Establishment and application of a point-of-care testing and diagnosis method for early immediate expression gene IE1 of cytomegalovirus in maternal urine based on isothermal amplification
Abstract
Background: Human Cytomegalovirus virus (HCMV) is a worldwide virus that causes no serious symptoms in most adults. However, HCMV infection during pregnancy, it may lead to a series of serious complications, such as hearing loss, mental retardation, visual impairment, microcephaly and developmental retardation.
Aim: The aim of this study was to develop a simple, low dependence on equipment and accurate method for HCMV detection based on the recombinase polymerase amplification (RPA) and lateral flow chromatography strip (LFS) reading.
Methods: In order to meet the feasibility of HCMV early screening, three pairs of RPA primers were designed based on the UL123 gene encoding IE1, which was expressed immediately in the early stage of HCMV. In order to improve the specificity of the reaction and satisfy the visual detection, a specific probe was designed to insert THF site between upstream and downstream primers, fluorescein isothiocyanate (FITC) and C3spacer were used to modify the 5' end and the 3' end respectively, and Biotin was used to modify the 5' end of the reverse primer. HCMV standard strain AD169 was enriched by ARPE-19 cells culture, and its genome was extracted. The primers and probes were screened by RPA-LFS test, and the optimal reaction temperature and time were determined The specificity was verified in different viruses, bacteria and parasites. The standard curve was drawn based on the constructed recombinant plasmid of pMD18T-HCMV-UL123 and used for HCMV genomic DNA quantification and determination of the detection sensitivity. Urine samples from artificial HCMV contamination or clinical collection were prepared to evaluate the consistency with the results of real-time quantitative PCR.
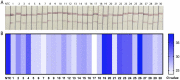
Results: The results showed that the primers and probes for HCMV RPA-LFS detection based on UL123 gene were successfully screened, the amplification of HCMV genomic DNA with as low as 30 copies could be completed at 37 °C within 15 min, it did not react with Human herpesvirus 1, Streptococcus pyogenes, Candida albicans, Listeria monocytogenes, Y. enterocolitica, Klebsiella Pneumoniae, Enterobacter cloacae, Citrobacter freundii, Vibrio alginnolyfificus, Vibrio parahaemolyticus, S. typhimurium, Staphylococcus aureus, Pseudomonas aeruginosa and Trichomonas vaginalis. The positive rate of PCR was 96.67 % in 30 simulated urine samples and 100 % in 127 clinical urine samples with the same UL123 gene detection.
Conclusions: To sum up, we developed a diagnostic method for HCMV based on UL123 gene combined with RPA and LFS, which is low dependent on equipment, fast, sensitive and specific, provide reference for point-of-care testing HCMV in grass-roots laboratories and remote areas.
Keywords: Early immediate expression gene; Human cytomegalovirus; Isothermal amplification; Lateral flow chromatography strip; THF.
Copyright © 2023. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
Establishment and application of a rapid molecular diagnostic platform for the isothermal visual amplification of group B Streptococcus based on recombinase polymerase.Front Cell Infect Microbiol. 2024 Feb 23;14:1281827. doi: 10.3389/fcimb.2024.1281827. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38465235 Free PMC article.
-
An improved recombinase polymerase amplification assay for visual detection of Vibrio parahaemolyticus with lateral flow strips.J Food Sci. 2020 Jun;85(6):1834-1844. doi: 10.1111/1750-3841.15105. Epub 2020 May 25. J Food Sci. 2020. PMID: 32449955
-
Rapid Detection of Klebsiella pneumoniae Carrying Virulence Gene rmpA2 by Recombinase Polymerase Amplification Combined With Lateral Flow Strips.Front Cell Infect Microbiol. 2022 May 19;12:877649. doi: 10.3389/fcimb.2022.877649. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35663473 Free PMC article.
-
[Preliminary application of real-time fluorescence recombinase polymerase amplification in Candida albicans].Zhonghua Shao Shang Za Zhi. 2019 Aug 20;35(8):587-594. doi: 10.3760/cma.j.issn.1009-2587.2019.08.006. Zhonghua Shao Shang Za Zhi. 2019. PMID: 31474038 Chinese.
-
Recombinant polymerase amplification combined with lateral flow strips for the detection of deep-seated Candida krusei infections.Front Cell Infect Microbiol. 2022 Jul 29;12:958858. doi: 10.3389/fcimb.2022.958858. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36004333 Free PMC article.
References
-
- Simonnet A., Engelmann I., Moreau A.S., Garcia B., Six S., El Kalioubie A., Robriquet L., Hober D., Jourdain M. High incidence of Epstein-Barr virus, cytomegalovirus, and human-herpes virus-6 reactivations in critically ill patients with COVID-19. Infect. Dis. Now. 2021;51(3):296–299. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

