Effects of spike proteins on angiotensin converting enzyme 2 (ACE2)
- PMID: 37769892
- PMCID: PMC10615800
- DOI: 10.1016/j.abb.2023.109769
Effects of spike proteins on angiotensin converting enzyme 2 (ACE2)
Abstract
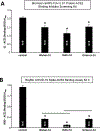
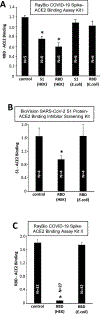
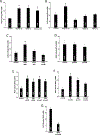
The Coronavirus Disease 2019 (COVID-19) pandemic was caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), which enters host cells through interactions of its spike protein to Angiotensin-Converting Enzyme 2 (ACE2). ACE2 is a peptidase that cleaves Angiotensin II, a critical pathological mediator. This study investigated if the spike protein binding to ACE2 compromises its peptidase activity. Spike/ACE2 Binding Assays suggested that spike proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, but not HKU1, bind to ACE2. S1 and receptor-binding domain (RBD), but not S2, extracellular domain (ECD) or CendR domain, bind to ACE2. While glycosylated spike proteins prepared in HEK293 cells bind to ACE2, non-glycosylated proteins produced in E. coli do not. Cysteine residues of the spike protein expressed in HEK293 cells are fully oxidized, while those of the protein expressed in E. coli are reduced. The deglycosylation of HEK cell-produced protein attenuates the ACE2 binding, while the oxidation of the E. coli protein does not promote the binding. The S1 protein of SARS-CoV-2 enhances the ACE2 peptidase activity, while SARS-CoV, MERS-CoV or HKU1 does not. The ACE2 activity is enhanced by RBD, but not ECD or CendR. In contrast to distinct ACE2 binding capacities of proteins expressed in HEK293 cells and in E. coli, spike proteins expressed in both systems enhance the ACE2 activity. Thus, the spike protein of SARS-CoV-2, but not other coronaviruses, enhances the ACE2 peptidase activity through its RBD in a glycosylation-independent manner.
Keywords: ACE2; COVID-19; Coronavirus; Peptidase; SARS-CoV-2; Spike protein.
Copyright © 2023 Elsevier Inc. All rights reserved.
Figures


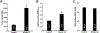
Similar articles
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
N-glycosylation of the SARS-CoV-2 spike protein at Asn331 and Asn343 is involved in spike-ACE2 binding, virus entry, and regulation of IL-6.Microbiol Immunol. 2024 May;68(5):165-178. doi: 10.1111/1348-0421.13121. Epub 2024 Mar 6. Microbiol Immunol. 2024. PMID: 38444370 Free PMC article.
-
Multidisciplinary Approaches Identify Compounds that Bind to Human ACE2 or SARS-CoV-2 Spike Protein as Candidates to Block SARS-CoV-2-ACE2 Receptor Interactions.mBio. 2021 Mar 30;12(2):e03681-20. doi: 10.1128/mBio.03681-20. mBio. 2021. PMID: 33785634 Free PMC article.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
-
Potential use of the S-protein-Angiotensin converting enzyme 2 binding pathway in the treatment of coronavirus disease 2019.Front Public Health. 2022 Nov 28;10:1050034. doi: 10.3389/fpubh.2022.1050034. eCollection 2022. Front Public Health. 2022. PMID: 36518573 Free PMC article. Review.
Cited by
-
Rice-derived SARS-CoV-2 glycoprotein S1 subunit vaccine elicits humoral and cellular immune responses.Plant Biotechnol J. 2025 Jul;23(7):2570-2582. doi: 10.1111/pbi.70077. Epub 2025 Apr 4. Plant Biotechnol J. 2025. PMID: 40183251 Free PMC article.
-
Characterizations of angiotensin-converting enzyme-2 (ACE2) peptidase activity.Arch Biochem Biophys. 2024 Nov;761:110167. doi: 10.1016/j.abb.2024.110167. Epub 2024 Sep 28. Arch Biochem Biophys. 2024. PMID: 39349131
-
Synthesis, molecular docking, and in vitro activity of a novel angiotensin-converting enzyme 2 inhibitor, LMS1007: a potential molecule in Covid-19 and cancer treatments.RSC Adv. 2025 May 8;15(19):15138-15154. doi: 10.1039/d5ra01134e. eCollection 2025 May 6. RSC Adv. 2025. PMID: 40343305 Free PMC article.
References
-
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet 395 (2020) 497–506. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

