Systematic Literature Review of Machine Learning Algorithms Using Pretherapy Radiologic Imaging for Glioma Molecular Subtype Prediction
- PMID: 37770204
- PMCID: PMC10549943
- DOI: 10.3174/ajnr.A8000
Systematic Literature Review of Machine Learning Algorithms Using Pretherapy Radiologic Imaging for Glioma Molecular Subtype Prediction
Abstract
Background: The molecular profile of gliomas is a prognostic indicator for survival, driving clinical decision-making for treatment. Pathology-based molecular diagnosis is challenging because of the invasiveness of the procedure, exclusion from neoadjuvant therapy options, and the heterogeneous nature of the tumor.
Purpose: We performed a systematic review of algorithms that predict molecular subtypes of gliomas from MR Imaging.
Data sources: Data sources were Ovid Embase, Ovid MEDLINE, Cochrane Central Register of Controlled Trials, Web of Science.
Study selection: Per the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines, 12,318 abstracts were screened and 1323 underwent full-text review, with 85 articles meeting the inclusion criteria.
Data analysis: We compared prediction results from different machine learning approaches for predicting molecular subtypes of gliomas. Bias analysis was conducted for each study, following the Prediction model Risk Of Bias Assessment Tool (PROBAST) guidelines.
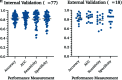
Data synthesis: Isocitrate dehydrogenase mutation status was reported with an area under the curve and accuracy of 0.88 and 85% in internal validation and 0.86 and 87% in limited external validation data sets, respectively. For the prediction of O6-methylguanine-DNA methyltransferase promoter methylation, the area under the curve and accuracy in internal validation data sets were 0.79 and 77%, and in limited external validation, 0.89 and 83%, respectively. PROBAST scoring demonstrated high bias in all articles.
Limitations: The low number of external validation and studies with incomplete data resulted in unequal data analysis. Comparing the best prediction pipelines of each study may introduce bias.
Conclusions: While the high area under the curve and accuracy for the prediction of molecular subtypes of gliomas are reported in internal and external validation data sets, limited use of external validation and the increased risk of bias in all articles may present obstacles for clinical translation of these techniques.
© 2023 by American Journal of Neuroradiology.
Figures





References
-
- Johns Hopkins Medicine. Gliomas. June 27, 2022. https://www.hopkinsmedicine.org/health/conditions-and-diseases/gliomas. Accessed June 27, 2022
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
