Defining the condensate landscape of fusion oncoproteins
- PMID: 37770423
- PMCID: PMC10539325
- DOI: 10.1038/s41467-023-41655-2
Defining the condensate landscape of fusion oncoproteins
Abstract
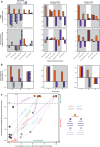
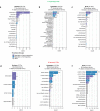
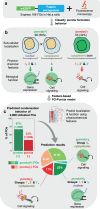
Fusion oncoproteins (FOs) arise from chromosomal translocations in ~17% of cancers and are often oncogenic drivers. Although some FOs can promote oncogenesis by undergoing liquid-liquid phase separation (LLPS) to form aberrant biomolecular condensates, the generality of this phenomenon is unknown. We explored this question by testing 166 FOs in HeLa cells and found that 58% formed condensates. The condensate-forming FOs displayed physicochemical features distinct from those of condensate-negative FOs and segregated into distinct feature-based groups that aligned with their sub-cellular localization and biological function. Using Machine Learning, we developed a predictor of FO condensation behavior, and discovered that 67% of ~3000 additional FOs likely form condensates, with 35% of those predicted to function by altering gene expression. 47% of the predicted condensate-negative FOs were associated with cell signaling functions, suggesting a functional dichotomy between condensate-positive and -negative FOs. Our Datasets and reagents are rich resources to interrogate FO condensation in the future.
© 2023. Springer Nature Limited.
Conflict of interest statement
S.D.G. is currently employed by Arrakis Therapeutics but his authorship role occurred while he was employed at St. Jude Children’s Research Hospital (SJCRH). I.I. has received honoraria from Amgen and Mission Bio. D.M.M. is currently employed by Dewpoint Therapeutics but her authorship role occurred while she was employed at SJCRH. M.R.W. is currently employed by IDEXX Laboratories, Inc. but his authorship role occurred while he was employed at SJCRH. D.F.J. reports personal fees from Transition Bio outside the submitted work. C.G.M. has received consulting and speaking fees from Illumina and Amgen, and research support from Loxo Oncology, Pfizer and Abbvie. R.W.K. reports personal fees from Dewpoint Therapeutics, GLG Consulting, and New Equilibrium Biosciences outside the submitted work. No disclosures were reported by the other authors.
Figures



References
-
- Brien GL, Stegmaier K, Armstrong SA. Targeting chromatin complexes in fusion protein-driven malignancies. Nat. Rev. Cancer. 2019;19:255–269. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

