Acetylation reprograms MITF target selectivity and residence time
- PMID: 37770430
- PMCID: PMC10539308
- DOI: 10.1038/s41467-023-41793-7
Acetylation reprograms MITF target selectivity and residence time
Abstract
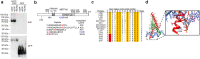
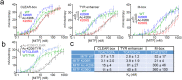
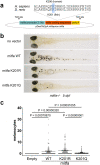
The ability of transcription factors to discriminate between different classes of binding sites associated with specific biological functions underpins effective gene regulation in development and homeostasis. How this is achieved is poorly understood. The microphthalmia-associated transcription factor MITF is a lineage-survival oncogene that plays a crucial role in melanocyte development and melanoma. MITF suppresses invasion, reprograms metabolism and promotes both proliferation and differentiation. How MITF distinguishes between differentiation and proliferation-associated targets is unknown. Here we show that compared to many transcription factors MITF exhibits a very long residence time which is reduced by p300/CBP-mediated MITF acetylation at K206. While K206 acetylation also decreases genome-wide MITF DNA-binding affinity, it preferentially directs DNA binding away from differentiation-associated CATGTG motifs toward CACGTG elements. The results reveal an acetylation-mediated switch that suppresses differentiation and provides a mechanistic explanation of why a human K206Q MITF mutation is associated with Waardenburg syndrome.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests
Figures



References
-
- Klemm SL, Shipony Z, Greenleaf WJ. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019;20:207–220. - PubMed
-
- Hodgkinson CA, et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

