Perivascular niche cells sense thrombocytopenia and activate hematopoietic stem cells in an IL-1 dependent manner
- PMID: 37770432
- PMCID: PMC10539537
- DOI: 10.1038/s41467-023-41691-y
Perivascular niche cells sense thrombocytopenia and activate hematopoietic stem cells in an IL-1 dependent manner
Abstract
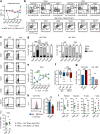
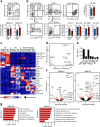
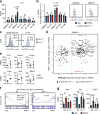
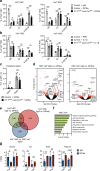
Hematopoietic stem cells (HSCs) residing in specialized niches in the bone marrow are responsible for the balanced output of multiple short-lived blood cell lineages in steady-state and in response to different challenges. However, feedback mechanisms by which HSCs, through their niches, sense acute losses of specific blood cell lineages remain to be established. While all HSCs replenish platelets, previous studies have shown that a large fraction of HSCs are molecularly primed for the megakaryocyte-platelet lineage and are rapidly recruited into proliferation upon platelet depletion. Platelets normally turnover in an activation-dependent manner, herein mimicked by antibodies inducing platelet activation and depletion. Antibody-mediated platelet activation upregulates expression of Interleukin-1 (IL-1) in platelets, and in bone marrow extracellular fluid in vivo. Genetic experiments demonstrate that rather than IL-1 directly activating HSCs, activation of bone marrow Lepr+ perivascular niche cells expressing IL-1 receptor is critical for the optimal activation of quiescent HSCs upon platelet activation and depletion. These findings identify a feedback mechanism by which activation-induced depletion of a mature blood cell lineage leads to a niche-dependent activation of HSCs to reinstate its homeostasis.
© 2023. The Author(s).
Conflict of interest statement
H.N. is the President and CEO, and G.Z. is an employee of CCOA Therapeutics Inc. Both authors are inventors of the monoclonal antibody NIT E (Canada patent number: 2, 689, 726; US patent number: US8, 323, 652 B2; and European patent number: 2186829). All other authors declare no competing interests.
Figures


References
-
- Takizawa H, Boettcher S, Manz MG. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 2012;119:2991–3002. - PubMed
-
- Wilson A, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–1129. - PubMed
-
- Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

