Multi-ancestry genome-wide study identifies effector genes and druggable pathways for coronary artery calcification
- PMID: 37770635
- PMCID: PMC10601987
- DOI: 10.1038/s41588-023-01518-4
Multi-ancestry genome-wide study identifies effector genes and druggable pathways for coronary artery calcification
Abstract
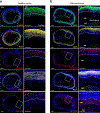
Coronary artery calcification (CAC), a measure of subclinical atherosclerosis, predicts future symptomatic coronary artery disease (CAD). Identifying genetic risk factors for CAC may point to new therapeutic avenues for prevention. Currently, there are only four known risk loci for CAC identified from genome-wide association studies (GWAS) in the general population. Here we conducted the largest multi-ancestry GWAS meta-analysis of CAC to date, which comprised 26,909 individuals of European ancestry and 8,867 individuals of African ancestry. We identified 11 independent risk loci, of which eight were new for CAC and five had not been reported for CAD. These new CAC loci are related to bone mineralization, phosphate catabolism and hormone metabolic pathways. Several new loci harbor candidate causal genes supported by multiple lines of functional evidence and are regulators of smooth muscle cell-mediated calcification ex vivo and in vitro. Together, these findings help refine the genetic architecture of CAC and extend our understanding of the biological and potential druggable pathways underlying CAC.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
S.W.v.d.L. has received Roche funding for unrelated work. B.M.P. serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. R.M. receives research funding from Angea Biotherapeutics and Amgen and serves as a consultant for Myokardia/BMS, Renovacor, Epizon Pharma, and Third Pole, all unrelated to the current project. C.L.M. has received funding from AstraZeneca on an unrelated project. J.C.K. is the recipient of an Agilent Thought Leader Award (January 2022), which includes funding for research that is unrelated to the current manuscript. All other co-authors confirm that they have no conflicts of interest related to the presented work.
Figures


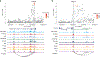



References
-
- Timmis A et al. European society of cardiology: Cardiovascular disease statistics 2017. Eur. Heart J. 39, 508–579 (2018). - PubMed
-
- Tsao CW et al. Heart Disease and Stroke Statistics-2022 Update: A Report From the American Heart Association. Circulation 145, e153–e639 (2022). - PubMed
-
- Libby P, Ridker PM & Hansson GK Progress and challenges in translating the biology of atherosclerosis. Nature 473, 317–325 (2011). - PubMed
-
- Baber U et al. Prevalence, impact, and predictive value of detecting subclinical coronary and carotid atherosclerosis in asymptomatic adults: the BioImage study. J. Am. Coll. Cardiol. 65, 1065–1074 (2015). - PubMed
Methods-only References
Publication types
MeSH terms
Grants and funding
- R01 HL125863/HL/NHLBI NIH HHS/United States
- P30 DK048520/DK/NIDDK NIH HHS/United States
- R01 HL162928/HL/NHLBI NIH HHS/United States
- F31 HL156463/HL/NHLBI NIH HHS/United States
- R01 HL163972/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- R01 HL159514/HL/NHLBI NIH HHS/United States
- R01 HL148239/HL/NHLBI NIH HHS/United States
- R01 HL142809/HL/NHLBI NIH HHS/United States
- K01 HL164687/HL/NHLBI NIH HHS/United States
- P30 DK063491/DK/NIDDK NIH HHS/United States
- R01 HL146860/HL/NHLBI NIH HHS/United States
- R01 DK114183/DK/NIDDK NIH HHS/United States
- R01 HL164577/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

