Distinct chemical environments in biomolecular condensates
- PMID: 37770698
- PMCID: PMC12181805
- DOI: 10.1038/s41589-023-01432-0
Distinct chemical environments in biomolecular condensates
Erratum in
-
Author Correction: Distinct chemical environments in biomolecular condensates.Nat Chem Biol. 2023 Dec;19(12):1561. doi: 10.1038/s41589-023-01491-3. Nat Chem Biol. 2023. PMID: 37880420 No abstract available.
Abstract
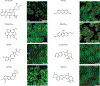
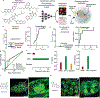
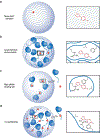
Diverse mechanisms have been described for selective enrichment of biomolecules in membrane-bound organelles, but less is known about mechanisms by which molecules are selectively incorporated into biomolecular assemblies such as condensates that lack surrounding membranes. The chemical environments within condensates may differ from those outside these bodies, and if these differed among various types of condensate, then the different solvation environments would provide a mechanism for selective distribution among these intracellular bodies. Here we use small molecule probes to show that different condensates have distinct chemical solvating properties and that selective partitioning of probes in condensates can be predicted with deep learning approaches. Our results demonstrate that different condensates harbor distinct chemical environments that influence the distribution of molecules, show that clues to condensate chemical grammar can be ascertained by machine learning and suggest approaches to facilitate development of small molecule therapeutics with optimal subcellular distribution and therapeutic benefit.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests:
R.A.Y. is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics, Dewpoint Therapeutics and Paratus Sciences, and has consulting or advisory roles at Precede Biosciences and Novo Nordisk. R.B. has consulting or advisory roles at Dewpoint Therapeutics, J&J, Amgen, Outcomes4Me, Immunai, and Firmenich. H.R.K. is a consultant of Dewpoint Therapeutics. The remaining authors declare no competing interests.
Figures






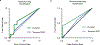




Comment in
-
Come for the atmosphere, stay for the interactions: Deciphering small molecule partitioning into biomolecular condensates.Cell Chem Biol. 2023 Nov 16;30(11):1337-1339. doi: 10.1016/j.chembiol.2023.10.015. Cell Chem Biol. 2023. PMID: 37977129
References
-
- Alberts B et al. Molecular biology of the cell. 7th edn, (W. W. Norton & Company, 2022).
-
- Shin Y & Brangwynne CP Liquid phase condensation in cell physiology and disease. Science 357 (2017). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

