Genomic epidemiology reveals multidrug resistant plasmid spread between Vibrio cholerae lineages in Yemen
- PMID: 37770747
- PMCID: PMC10539172
- DOI: 10.1038/s41564-023-01472-1
Genomic epidemiology reveals multidrug resistant plasmid spread between Vibrio cholerae lineages in Yemen
Abstract
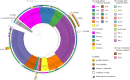
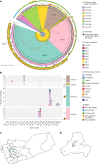
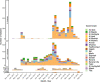
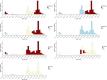
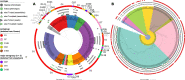
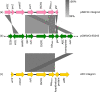
Since 2016, Yemen has been experiencing the largest cholera outbreak in modern history. Multidrug resistance (MDR) emerged among Vibrio cholerae isolates from cholera patients in 2018. Here, to characterize circulating genotypes, we analysed 260 isolates sampled in Yemen between 2018 and 2019. Eighty-four percent of V. cholerae isolates were serogroup O1 belonging to the seventh pandemic El Tor (7PET) lineage, sub-lineage T13, whereas 16% were non-toxigenic, from divergent non-7PET lineages. Treatment of severe cholera with macrolides between 2016 and 2019 coincided with the emergence and dominance of T13 subclones carrying an incompatibility type C (IncC) plasmid harbouring an MDR pseudo-compound transposon. MDR plasmid detection also in endemic non-7PET V. cholerae lineages suggested genetic exchange with 7PET epidemic strains. Stable co-occurrence of the IncC plasmid with the SXT family of integrative and conjugative element in the 7PET background has major implications for cholera control, highlighting the importance of genomic epidemiological surveillance to limit MDR spread.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

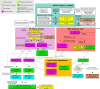
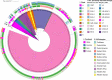
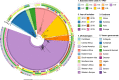
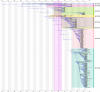
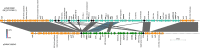

References
-
- UNHCR Yemen: 2021 Country Operational Plan (UNHCR, 2021); https://data2.unhcr.org/en/documents/details/85850
-
- Cholera (WHO EMRO, 2021); http://www.emro.who.int/health-topics/cholera-outbreak/cholera-outbreaks...
-
- Health workers in Yemen reach more than 306,000 people with cholera vaccines during four-day pause in fighting—WHO, UNICEF. WHOhttps://www.who.int/news/item/05-10-2018-health-workers-in-yemen-reach-m... (2018).
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

