Phylodynamics of dengue virus 2 in Nicaragua leading up to the 2019 epidemic reveals a role for lineage turnover
- PMID: 37770825
- PMCID: PMC10537812
- DOI: 10.1186/s12862-023-02156-4
Phylodynamics of dengue virus 2 in Nicaragua leading up to the 2019 epidemic reveals a role for lineage turnover
Abstract
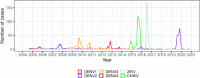
Background: Dengue is a mosquito-borne viral disease posing a significant threat to public health. Dengue virus (DENV) evolution is often characterized by lineage turnover, which, along with ecological and immunological factors, has been linked to changes in dengue phenotype affecting epidemic dynamics. Utilizing epidemiologic and virologic data from long-term population-based studies (the Nicaraguan Pediatric Dengue Cohort Study and Nicaraguan Dengue Hospital-based Study), we describe a lineage turnover of DENV serotype 2 (DENV-2) prior to a large dengue epidemic in 2019. Prior to this epidemic, Nicaragua had experienced relatively low levels of DENV transmission from 2014 to 2019, a period dominated by chikungunya in 2014/15 and Zika in 2016.
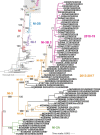
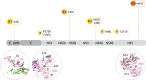
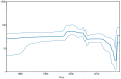
Results: Our phylogenetic analyses confirmed that all Nicaraguan DENV-2 isolates from 2018 to 2019 formed their own clade within the Nicaraguan lineage of the Asian/American genotype. The emergence of the new DENV-2 lineage reflects a replacement of the formerly dominant clade presiding from 2005 to 2009, a lineage turnover marked by several shared derived amino acid substitutions throughout the genome. To elucidate evolutionary drivers of lineage turnover, we performed selection pressure analysis and reconstructed the demographic history of DENV-2. We found evidence of adaptive evolution by natural selection at the codon level as well as in branch formation.
Conclusions: The timing of its emergence, along with a statistical signal of adaptive evolution and distinctive amino acid substitutions, the latest in the NS5 gene, suggest that this lineage may have increased fitness relative to the prior dominant DENV-2 strains. This may have contributed to the intensity of the 2019 DENV-2 epidemic, in addition to previously identified immunological factors associated with pre-existing Zika virus immunity.
Keywords: Clade replacement; Dengue virus; Lineage turnover; Natural selection; Nicaragua; Phylodynamics; Virus evolution.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Introduction of New Dengue Virus Lineages of Multiple Serotypes after COVID-19 Pandemic, Nicaragua, 2022.Emerg Infect Dis. 2024 Jun;30(6):1203-1213. doi: 10.3201/eid3006.231553. Emerg Infect Dis. 2024. PMID: 38782023 Free PMC article.
-
Evolution and epidemiologic dynamics of dengue virus in Nicaragua during the emergence of chikungunya and Zika viruses.Infect Genet Evol. 2021 Aug;92:104680. doi: 10.1016/j.meegid.2020.104680. Epub 2020 Dec 14. Infect Genet Evol. 2021. PMID: 33326875 Free PMC article.
-
Prior dengue virus infection and risk of Zika: A pediatric cohort in Nicaragua.PLoS Med. 2019 Jan 22;16(1):e1002726. doi: 10.1371/journal.pmed.1002726. eCollection 2019 Jan. PLoS Med. 2019. PMID: 30668565 Free PMC article.
-
Potential role of vector-mediated natural selection in dengue virus genotype/lineage replacements in two epidemiologically contrasted settings.Emerg Microbes Infect. 2021 Dec;10(1):1346-1357. doi: 10.1080/22221751.2021.1944789. Emerg Microbes Infect. 2021. PMID: 34139961 Free PMC article.
-
Genetics of dengue epidemics.Trends Microbiol. 2025 Jun 5:S0966-842X(25)00154-4. doi: 10.1016/j.tim.2025.05.007. Online ahead of print. Trends Microbiol. 2025. PMID: 40480856 Review.
Cited by
-
Serotype-specific epidemiological patterns of inapparent versus symptomatic primary dengue virus infections: a 17-year cohort study in Nicaragua.Lancet Infect Dis. 2025 Mar;25(3):346-356. doi: 10.1016/S1473-3099(24)00566-8. Epub 2024 Oct 25. Lancet Infect Dis. 2025. PMID: 39489898 Free PMC article.
-
A new lineage nomenclature to aid genomic surveillance of dengue virus.PLoS Biol. 2024 Sep 16;22(9):e3002834. doi: 10.1371/journal.pbio.3002834. eCollection 2024 Sep. PLoS Biol. 2024. PMID: 39283942 Free PMC article.
-
Introduction of New Dengue Virus Lineages of Multiple Serotypes after COVID-19 Pandemic, Nicaragua, 2022.Emerg Infect Dis. 2024 Jun;30(6):1203-1213. doi: 10.3201/eid3006.231553. Emerg Infect Dis. 2024. PMID: 38782023 Free PMC article.
-
A new lineage nomenclature to aid genomic surveillance of dengue virus.medRxiv [Preprint]. 2024 May 17:2024.05.16.24307504. doi: 10.1101/2024.05.16.24307504. medRxiv. 2024. Update in: PLoS Biol. 2024 Sep 16;22(9):e3002834. doi: 10.1371/journal.pbio.3002834. PMID: 38798319 Free PMC article. Updated. Preprint.
-
Spatiotemporal dispersion of DENV-1 genotype V in Western Colombia.Virus Evol. 2025 Apr 16;11(1):veaf018. doi: 10.1093/ve/veaf018. eCollection 2025. Virus Evol. 2025. PMID: 40395613 Free PMC article.
References
-
- Gubler DJ, Ooi EE, Vasudevan S, Farrar J. Dengue and Dengue Hemorrhagic Fever, 2nd Edition. CABI; 2014.
-
- WHO. | Dengue and severe dengue. 2017.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
