Genotypic and phenotypic comparison of drug resistance profiles of clinical multidrug-resistant Mycobacterium tuberculosis isolates using whole genome sequencing in Latvia
- PMID: 37770850
- PMCID: PMC10540372
- DOI: 10.1186/s12879-023-08629-7
Genotypic and phenotypic comparison of drug resistance profiles of clinical multidrug-resistant Mycobacterium tuberculosis isolates using whole genome sequencing in Latvia
Abstract
Background: Multidrug-resistant tuberculosis (MDR-TB) remains a major public health problem in many high tuberculosis (TB) burden countries. Phenotypic drug susceptibility testing (DST) take several weeks or months to result, but line probe assays and Xpert/Rif Ultra assay detect a limited number of resistance conferring gene mutations. Whole genome sequencing (WGS) is an advanced molecular testing method which theoretically can predict the resistance of M. tuberculosis (Mtb) isolates to all anti-TB agents through a single analysis.
Methods: Here, we aimed to identify the level of concordance between the phenotypic and WGS-based genotypic drug susceptibility (DS) patterns of MDR-TB isolates. Overall, data for 12 anti-TB medications were analyzed.
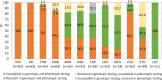
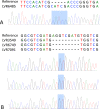
Results: In total, 63 MDR-TB Mtb isolates were included in the analysis, representing 27.4% of the total number of MDR-TB cases in Latvia in 2012-2014. Among them, five different sublineages were detected, and 2.2.1 (Beijing group) and 4.3.3 (Latin American-Mediterranean group) were the most abundant. There were 100% agreement between phenotypic and genotypic DS pattern for isoniazid, rifampicin, and linezolid. High concordance rate (> 90%) between phenotypic and genotypic DST results was detected for ofloxacin (93.7%), pyrazinamide (93.7%) and streptomycin (95.4%). Phenotypic and genotypic DS patterns were poorly correlated for ethionamide (agreement 56.4%), ethambutol (85.7%), amikacin (82.5%), capreomycin (81.0%), kanamycin (85.4%), and moxifloxacin (77.8%). For capreomycin, resistance conferring mutations were not identified in several phenotypically resistant isolates, and, in contrary, for ethionamide, ethambutol, amikacin, kanamycin, and moxifloxacin the resistance-related mutations were identified in several phenotypically sensitive isolates.
Conclusions: WGS is a valuable tool for rapid genotypic DST for all anti-TB agents. For isoniazid and rifampicin phenotypic DST potentially can be replaced by genotypic DST based on 100% agreement between the tests. However, discrepant results for other anti-TB agents limit their prescription based solely on WGS data. For clinical decision, at the current level of knowledge, there is a need for combination of genotypic DST with modern, validated phenotypic DST methodologies for those medications which did not showed 100% agreement between the methods.
Keywords: Drug susceptibility testing; Drug-resistant tuberculosis; Latvia; Whole genome sequencing.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Global tuberculosis report 2022 . Licence: CC BY-NC-SA 3.0 IGO. Geneva: World Health Organization; 2022.
-
- Ko DH, Lee EJ, Lee SK, Kim HS, Shin SY, Hyun J, Kim JS, Song W, Kim HS. Application of next-generation sequencing to detect variants of drug-resistant Mycobacterium tuberculosis: genotype-phenotype correlation. Ann Clin Microbiol Antimicrob. 2019;18(1):2. doi: 10.1186/s12941-018-0300-y. - DOI - PMC - PubMed
-
- Walker TM, Kohl TA, Omar SV, Hedge J, Del Ojo Elias C, Bradley P, Iqbal Z, Feuerriegel S, Niehaus KE, Wilson DJ, Clifton DA, Kapatai G, Ip CLC, Bowden R, Drobniewski FA, Allix-Béguec C, Gaudin C, Parkhill J, Diel R, Supply P, Crook DW, Smith EG, Walker AS, Ismail N, Niemann S, Peto TEA. Modernizing Medical Microbiology (MMM) Informatics Group. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis. 2015;15(10):1193–202. doi: 10.1016/S1473-3099(15)00062-6. - DOI - PMC - PubMed
-
- Sun L, Zhang L, Wang T, Jiao W, Li Q, Yin Q, Li J, Qi H, Xu F, Shen C, Xiao J, Liu S, Mokrousov I, Huang H, Shen A. Mutations of Mycobacterium tuberculosis induced by anti-tuberculosis treatment result in metabolism changes and elevation of ethambutol resistance. Infect Genet Evol. 2019;72:151–8. doi: 10.1016/j.meegid.2018.09.027. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

