Network module analysis and molecular docking-based study on the mechanism of astragali radix against non-small cell lung cancer
- PMID: 37770919
- PMCID: PMC10537544
- DOI: 10.1186/s12906-023-04148-9
Network module analysis and molecular docking-based study on the mechanism of astragali radix against non-small cell lung cancer
Abstract
Background: Most lung cancer patients worldwide (stage IV non-small cell lung cancer, NSCLC) have a poor survival: 25%-30% patients die < 3 months. Yet, of those surviving > 3 months, 10%-15% patients survive (very) long. Astragali radix (AR) is an effective traditional Chinese medicine widely used for non-small cell lung cancer (NSCLC). However, the pharmacological mechanisms of AR on NSCLC remain to be elucidated.
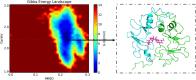
Methods: Ultra Performance Liquid Chromatography system coupled with Q-Orbitrap HRMS (UPLC-Q-Orbitrap HRMS) was performed for the qualitative analysis of AR components. Then, network module analysis and molecular docking-based approach was conducted to explore underlying mechanisms of AR on NSCLC. The target genes of AR were obtained from four databases including TCMSP (Traditional Chinese Medicine Systems Pharmacology) database, ETCM (The Encyclopedia of TCM) database, HERB (A high-throughput experiment- and reference-guided database of TCM) database and BATMAN-TCM (a Bioinformatics Analysis Tool for Molecular mechanism of TCM) database. NSCLC related genes were screened by GEO (Gene Expression Omnibus) database. The STRING database was used for protein interaction network construction (PIN) of AR-NSCLC shared target genes. The critical PIN were further constructed based on the topological properties of network nodes. Afterwards the hub genes and network modules were analyzed, and enrichment analysis were employed by the R package clusterProfiler. The Autodock Vina was utilized for molecular docking, and the Gromacs was utilized for molecular dynamics simulations Furthermore, the survival analysis was performed based on TCGA (The Cancer Genome Atlas) database.
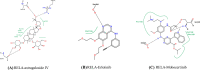
Results: Seventy-seven AR components absorbed in blood were obtained. The critical network was constructed with 1447 nodes and 28,890 edges. Based on topological analysis, 6 hub target genes and 7 functional modules were gained. were obtained including TP53, SRC, UBC, CTNNB1, EP300, and RELA. After module analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that AR may exert therapeutic effects on NSCLC by regulating JAK-STAT signaling pathway, PI3K-AKT signaling pathway, ErbB signaling pathway, as well as NFkB signaling pathway. After the intersection calculation of the hub targets and the proteins participated in the above pathways, TP53, SRC, EP300, and RELA were obtained. These proteins had good docking affinity with astragaloside IV. Furthermore, RELA was associated with poor prognosis of NSCLC patients.
Conclusions: This study could provide chemical component information references for further researches. The potential pharmacological mechanisms of AR on NSCLC were elucidated, promoting the clinical application of AR in treating NSCLC. RELA was selected as a promising candidate biomarker affecting the prognosis of NSCLC patients.
Keywords: Astragali radix; Molecular docking; Network pharmacology; Non-small cell lung cancer; TCGA.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Wu C, Song W, Wang Z, et al. Functions of lncRNA DUXAP8 in non-small cell lung cancer. Mol Biol Rep. 2022;49(3):2531–2542. - PubMed
-
- Pompili C, Koller M, Velikova G, et al. EORTC QLQ-C30 summary score reliably detects changes in QoL three months after anatomic lung resection for non-small cell lung cancer (NSCLC)[J]. Lung Cancer. 2018;123:149–54. - PubMed
-
- Rauma V, Sintonen H, Räsänen J V, et al. Long-term lung cancer survivors have permanently decreased quality of life after surgery[J]. Clin Lung Cancer. 2015;16(1):40–5. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

