Therapeutic targeting of the TPX2/TTK network in colorectal cancer
- PMID: 37770979
- PMCID: PMC10536736
- DOI: 10.1186/s12964-023-01290-2
Therapeutic targeting of the TPX2/TTK network in colorectal cancer
Abstract
Background: While the increased screening, changes in lifestyle, and recent advances in treatment regimen have decreased colorectal cancer (CRC) mortality, metastatic disease and recurrence remains a major clinical challenge. In the era of precision medicine, the identification of actionable novel therapeutic targets could ultimately offer an alternative treatment strategy for CRC.
Methods: RNA-Seq was conducted using the illumina platform, while bioinformatics analyses were conducted using CLC genomics workbench and iDEP.951. Colony forming unit, flow cytometry, and fluorescent microscopy were used to assess cell proliferation, cell cycle distribution, and cell death, respectively. The growth potential of CRC cells under 3-dimensional (3D) conditions was assessed using Matrigel. STRING database (v11.5) and Ingenuity Pathway Analysis (IPA) tool were used for network and pathway analyses. CRISPR-Cas9 perturbational effects database was used to identify potential therapeutic targets for CRC, through integration with gene-drug interaction database. Structural modeling and molecular docking were used to assess the interaction between candidate drugs and their targets.
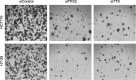
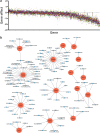
Results: In the current study, we investigated the therapeutic potential of targeting TPX2, TTK, DDX39A, and LRP8, commonly upregulated genes in CRC identified through differential expression analysis in CRC and adjacent non-cancerous tissue. Targeted depletion of TPX2 and TTK impaired CRC proliferation, cell cycle progression, and organoid formation under 3D culture conditions, while suppression of DDX39A and LRP8 had modest effects on CRC colony formation. Differential expression analysis and bioinformatics on TPX2 and TTK-deficient cells identified cell cycle regulation as the hallmark associated with loss of TPX2 and TTK. Elevated expression of TPX2 and TTK correlated with an oncogenic state in tumor tissue from patients with colon adenocarcinoma, thus corroborating an oncogenic role for the TPX2/TTK network in the pathogenesis of CRC. Gene set enrichment and pathway analysis of TPX2high/TTKhigh CRC identified numerous additional gene targets as integral components of the TPX2/TTK network. Integration of TPX2/TTK enriched network with CRISPR-Cas9 functional screen data identified numerous novel dependencies for CRC. Additionally, gene-drug interaction analysis identified several druggable gene targets enriched in the TPX2/TTK network, including AURKA, TOP2A, CDK1, BIRC5, and many others.
Conclusions: Our data has implicated an essential role for TPX2 and TTK in CRC pathogenesis and identified numerous potential therapeutic targets and their drug interactions, suggesting their potential clinical use as a novel therapeutic strategy for patients with CRC. Video Abstract.
Keywords: Colorectal cancer; DDX39A; Gene-drug-interaction; LRP8; Precision medicine; TPX2; TTK.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Davis LE. The evolution of biomarkers to guide the treatment of metastatic colorectal cancer. Am J Manag Care. 2018;24:S107–S117. - PubMed
-
- Van Cutsem E, Lenz HJ, Kohne CH, Heinemann V, Tejpar S, Melezinek I, Beier F, Stroh C, Rougier P, van Krieken JH, Ciardiello F. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol. 2015;33:692–700. - PubMed
-
- Elsafadi M, Manikandan M, Almalki S, Mahmood A, Shinwari T, Vishnubalaji R, Mobarak M, Alfayez M, Aldahmash A, Kassem M, Alajez NM. Transgelin is a poor prognostic factor associated with advanced colorectal cancer (CRC) stage promoting tumor growth and migration in a TGFbeta-dependent manner. Cell Death Dis. 2020;11:341. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

