Real-time monitoring strategies for optimization of in vitro transcription and quality control of RNA
- PMID: 37771458
- PMCID: PMC10523567
- DOI: 10.3389/fmolb.2023.1229246
Real-time monitoring strategies for optimization of in vitro transcription and quality control of RNA
Abstract
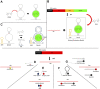
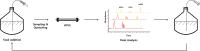
RNA-based therapeutics and vaccines are opening up new avenues for modern medicine. To produce these useful RNA-based reagents, in vitro transcription (IVT) is an important reaction that primarily determines the yield and quality of the product. Therefore, IVT condition should be well optimized to achieve high yield and purity of transcribed RNAs. To this end, real-time monitoring of RNA production during IVT, which allows for fine tuning of the condition, would be required. Currently, light-up RNA aptamer and fluorescent dye pairs are considered as useful strategies to monitor IVT in real time. Fluorophore-labeled antisense probe-based methods can also be used for real-time IVT monitoring. In addition, a high-performance liquid chromatography (HPLC)-based method that can monitor IVT reagent consumption has been developed as a powerful tool to monitor IVT reaction in near real-time. This mini-review briefly introduces some strategies and examples for real-time IVT monitoring and discusses pros and cons of IVT monitoring methods.
Keywords: IVT; RNA; in vitro transcription; real-time monitoring; transcription.
Copyright © 2023 Lee, Song, Kim, Han and Lee.
Conflict of interest statement
KHL, JS, SK, and SRH are employee of Rznomics Inc. S-WL is CEO of Rznomics Inc.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

