happi: a hierarchical approach to pangenomics inference
- PMID: 37773075
- PMCID: PMC10540326
- DOI: 10.1186/s13059-023-03040-6
happi: a hierarchical approach to pangenomics inference
Abstract
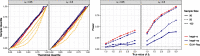
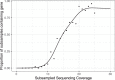
Recovering metagenome-assembled genomes (MAGs) from shotgun sequencing data is an increasingly common task in microbiome studies, as MAGs provide deeper insight into the functional potential of both culturable and non-culturable microorganisms. However, metagenome-assembled genomes vary in quality and may contain omissions and contamination. These errors present challenges for detecting genes and comparing gene enrichment across sample types. To address this, we propose happi, an approach to testing hypotheses about gene enrichment that accounts for genome quality. We illustrate the advantages of happi over existing approaches using published Saccharibacteria MAGs, Streptococcus thermophilus MAGs, and via simulation.
Keywords: Hypothesis testing; Metagenome-assembled genomes; Microbiome; Shotgun metagenomics; Statistical models.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Recovering metagenome-assembled genomes from shotgun metagenomic sequencing data: Methods, applications, challenges, and opportunities.Microbiol Res. 2022 Jul;260:127023. doi: 10.1016/j.micres.2022.127023. Epub 2022 Apr 8. Microbiol Res. 2022. PMID: 35430490 Review.
-
The Reliability of Metagenome-Assembled Genomes (MAGs) in Representing Natural Populations: Insights from Comparing MAGs against Isolate Genomes Derived from the Same Fecal Sample.Appl Environ Microbiol. 2021 Feb 26;87(6):e02593-20. doi: 10.1128/AEM.02593-20. Print 2021 Feb 26. Appl Environ Microbiol. 2021. PMID: 33452027 Free PMC article.
-
Long-read based de novo assembly of low-complexity metagenome samples results in finished genomes and reveals insights into strain diversity and an active phage system.BMC Microbiol. 2019 Jun 25;19(1):143. doi: 10.1186/s12866-019-1500-0. BMC Microbiol. 2019. PMID: 31238873 Free PMC article.
-
Novel canine high-quality metagenome-assembled genomes, prophages and host-associated plasmids provided by long-read metagenomics together with Hi-C proximity ligation.Microb Genom. 2022 Mar;8(3):000802. doi: 10.1099/mgen.0.000802. Microb Genom. 2022. PMID: 35298370 Free PMC article.
-
An Introduction to Whole-Metagenome Shotgun Sequencing Studies.Methods Mol Biol. 2021;2243:107-122. doi: 10.1007/978-1-0716-1103-6_6. Methods Mol Biol. 2021. PMID: 33606255 Review.
Cited by
-
Differences in gut metagenomes between dairy workers and community controls: a cross-sectional study.bioRxiv [Preprint]. 2023 May 12:2023.05.10.540270. doi: 10.1101/2023.05.10.540270. bioRxiv. 2023. Update in: BMC Genomics. 2024 Jul 20;25(1):708. doi: 10.1186/s12864-024-10562-1. PMID: 37215025 Free PMC article. Updated. Preprint.
-
A cross-sectional comparison of gut metagenomes between dairy workers and community controls.BMC Genomics. 2024 Jul 20;25(1):708. doi: 10.1186/s12864-024-10562-1. BMC Genomics. 2024. PMID: 39033279 Free PMC article.
-
Rapid species-level metagenome profiling and containment estimation with sylph.Nat Biotechnol. 2025 Aug;43(8):1348-1359. doi: 10.1038/s41587-024-02412-y. Epub 2024 Oct 8. Nat Biotechnol. 2025. PMID: 39379646 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

