TMEM135 links peroxisomes to the regulation of brown fat mitochondrial fission and energy homeostasis
- PMID: 37773161
- PMCID: PMC10541902
- DOI: 10.1038/s41467-023-41849-8
TMEM135 links peroxisomes to the regulation of brown fat mitochondrial fission and energy homeostasis
Abstract
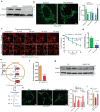
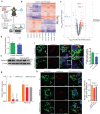
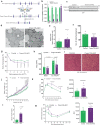
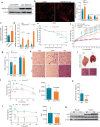
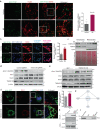
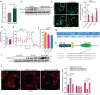
Mitochondrial morphology, which is controlled by mitochondrial fission and fusion, is an important regulator of the thermogenic capacity of brown adipocytes. Adipose-specific peroxisome deficiency impairs thermogenesis by inhibiting cold-induced mitochondrial fission due to decreased mitochondrial membrane content of the peroxisome-derived lipids called plasmalogens. Here, we identify TMEM135 as a critical mediator of the peroxisomal regulation of mitochondrial fission and thermogenesis. Adipose-specific TMEM135 knockout in mice blocks mitochondrial fission, impairs thermogenesis, and increases diet-induced obesity and insulin resistance. Conversely, TMEM135 overexpression promotes mitochondrial division, counteracts obesity and insulin resistance, and rescues thermogenesis in peroxisome-deficient mice. Mechanistically, thermogenic stimuli promote association between peroxisomes and mitochondria and plasmalogen-dependent localization of TMEM135 in mitochondria, where it mediates PKA-dependent phosphorylation and mitochondrial retention of the fission factor Drp1. Together, these results reveal a previously unrecognized inter-organelle communication regulating mitochondrial fission and energy homeostasis and identify TMEM135 as a potential target for therapeutic activation of BAT.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL125838/HL/NHLBI NIH HHS/United States
- R01 HL159461/HL/NHLBI NIH HHS/United States
- R01 DK102898/DK/NIDDK NIH HHS/United States
- R01 EY022086/EY/NEI NIH HHS/United States
- P30 DK056341/DK/NIDDK NIH HHS/United States
- P41 GM103422/GM/NIGMS NIH HHS/United States
- P30 DK036836/DK/NIDDK NIH HHS/United States
- R01 DK131188/DK/NIDDK NIH HHS/United States
- P30 CA091842/CA/NCI NIH HHS/United States
- P30 DK020579/DK/NIDDK NIH HHS/United States
- R01 DK118333/DK/NIDDK NIH HHS/United States
- R01 DK132469/DK/NIDDK NIH HHS/United States
- R01 DK133344/DK/NIDDK NIH HHS/United States
- R01 DK115867/DK/NIDDK NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- T32 DK007120/DK/NIDDK NIH HHS/United States
- P60 DK020579/DK/NIDDK NIH HHS/United States
- R01 DK132239/DK/NIDDK NIH HHS/United States
- S10 OD030403/OD/NIH HHS/United States
- R01 DK133528/DK/NIDDK NIH HHS/United States
- R24 GM136766/GM/NIGMS NIH HHS/United States
- UL1 TR002345/TR/NCATS NIH HHS/United States
- I01 BX003415/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

