Senescent fibro-adipogenic progenitors are potential drivers of pathology in inclusion body myositis
- PMID: 37773216
- PMCID: PMC10564677
- DOI: 10.1007/s00401-023-02637-2
Senescent fibro-adipogenic progenitors are potential drivers of pathology in inclusion body myositis
Abstract
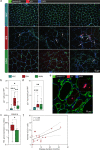
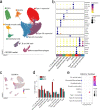
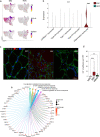
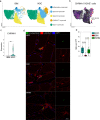
Inclusion body myositis (IBM) is unique across the spectrum of idiopathic inflammatory myopathies (IIM) due to its distinct clinical presentation and refractoriness to current treatment approaches. One explanation for this resistance may be the engagement of cell-autonomous mechanisms that sustain or promote disease progression of IBM independent of inflammatory activity. In this study, we focused on senescence of tissue-resident cells as potential driver of disease. For this purpose, we compared IBM patients to non-diseased controls and immune-mediated necrotizing myopathy patients. Histopathological analysis suggested that cellular senescence is a prominent feature of IBM, primarily affecting non-myogenic cells. In-depth analysis by single nuclei RNA sequencing allowed for the deconvolution and study of muscle-resident cell populations. Among these, we identified a specific cluster of fibro-adipogenic progenitors (FAPs) that demonstrated key hallmarks of senescence, including a pro-inflammatory secretome, expression of p21, increased β-galactosidase activity, and engagement of senescence pathways. FAP function is required for muscle cell health with changes to their phenotype potentially proving detrimental. In this respect, the transcriptomic landscape of IBM was also characterized by changes to the myogenic compartment demonstrating a pronounced loss of type 2A myofibers and a rarefication of acetylcholine receptor expressing myofibers. IBM muscle cells also engaged a specific pro-inflammatory phenotype defined by intracellular complement activity and the expression of immunogenic surface molecules. Skeletal muscle cell dysfunction may be linked to FAP senescence by a change in the collagen composition of the latter. Senescent FAPs lose collagen type XV expression, which is required to support myofibers' structural integrity and neuromuscular junction formation in vitro. Taken together, this study demonstrates an altered phenotypical landscape of muscle-resident cells and that FAPs, and not myofibers, are the primary senescent cell type in IBM.
Keywords: Acetylcholine receptor; Complement; Inclusion body myositis; Myofiber; Senescence; Single nuclei.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Allenbach Y, Mammen AL, Benveniste O, Stenzel W, Allenbach Y, Amato A, et al. 224th ENMC international workshop: clinico-sero-pathological classification of immune-mediated necrotizing myopathies Zandvoort, the Netherlands, 14–16 October 2016. Neuromuscul Disord. 2018;28:87–99. doi: 10.1016/j.nmd.2017.09.016. - DOI - PubMed
-
- Benveniste O, Hogrel J-Y, Belin L, Annoussamy M, Bachasson D, Rigolet A, et al. Sirolimus for treatment of patients with inclusion body myositis: a randomised, double-blind, placebo-controlled, proof-of-concept, phase 2b trial. Lancet Rheumatol. 2021;3:e40–e48. doi: 10.1016/S2665-9913(20)30280-0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

