Human retinal ganglion cell neurons generated by synchronous BMP inhibition and transcription factor mediated reprogramming
- PMID: 37773257
- PMCID: PMC10541876
- DOI: 10.1038/s41536-023-00327-x
Human retinal ganglion cell neurons generated by synchronous BMP inhibition and transcription factor mediated reprogramming
Abstract
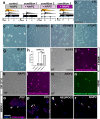
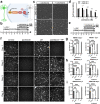
In optic neuropathies, including glaucoma, retinal ganglion cells (RGCs) die. Cell transplantation and endogenous regeneration offer strategies for retinal repair, however, developmental programs required for this to succeed are incompletely understood. To address this, we explored cellular reprogramming with transcription factor (TF) regulators of RGC development which were integrated into human pluripotent stem cells (PSCs) as inducible gene cassettes. When the pioneer factor NEUROG2 was combined with RGC-expressed TFs (ATOH7, ISL1, and POU4F2) some conversion was observed and when pre-patterned by BMP inhibition, RGC-like induced neurons (RGC-iNs) were generated with high efficiency in just under a week. These exhibited transcriptional profiles that were reminiscent of RGCs and exhibited electrophysiological properties, including AMPA-mediated synaptic transmission. Additionally, we demonstrated that small molecule inhibitors of DLK/LZK and GCK-IV can block neuronal death in two pharmacological axon injury models. Combining developmental patterning with RGC-specific TFs thus provided valuable insight into strategies for cell replacement and neuroprotection.
© 2023. Springer Nature Limited.
Conflict of interest statement
K.J.W., D.A., N.D., and R.M.W. have filed a United States provisional patent (63/478416) for work related to RGC reprogramming. J.M.R. is the chief medical officer for Path Therapeutics, Inc., consultant for Danone Nutricia, Cerecin, Eisai, Biocodex, Zogenix and in the speaker’s bureau for Aquestive Pharmaceuticals. The remaining authors declare no competing interests.
Figures





References
-
- Hasegawa Y, et al. Emergence of dorsal-ventral polarity in ESC-derived retinal tissue. Development. 2016;143:3895–3906. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

