Viral Membrane Fusion: A Dance Between Proteins and Lipids
- PMID: 37774128
- PMCID: PMC10866366
- DOI: 10.1146/annurev-virology-111821-093413
Viral Membrane Fusion: A Dance Between Proteins and Lipids
Abstract
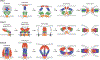
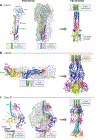
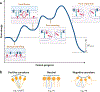
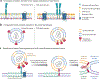
There are at least 21 families of enveloped viruses that infect mammals, and many contain members of high concern for global human health. All enveloped viruses have a dedicated fusion protein or fusion complex that enacts the critical genome-releasing membrane fusion event that is essential before viral replication within the host cell interior can begin. Because all enveloped viruses enter cells by fusion, it behooves us to know how viral fusion proteins function. Viral fusion proteins are also major targets of neutralizing antibodies, and hence they serve as key vaccine immunogens. Here we review current concepts about viral membrane fusion proteins focusing on how they are triggered, structural intermediates between pre- and postfusion forms, and their interplay with the lipid bilayers they engage. We also discuss cellular and therapeutic interventions that thwart virus-cell membrane fusion.
Keywords: class I viral fusion proteins; class II viral fusion proteins; class III viral fusion proteins; conformational intermediates; fusion energetics; fusion loops; fusion peptides; fusion restriction factors; lipid dynamics; trimers-of-hairpins.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

