Preclinical development of a chimeric antigen receptor T cell therapy targeting FGFR4 in rhabdomyosarcoma
- PMID: 37774704
- PMCID: PMC10591056
- DOI: 10.1016/j.xcrm.2023.101212
Preclinical development of a chimeric antigen receptor T cell therapy targeting FGFR4 in rhabdomyosarcoma
Erratum in
-
Preclinical development of a chimeric antigen receptor T cell therapy targeting FGFR4 in rhabdomyosarcoma.Cell Rep Med. 2024 May 21;5(5):101586. doi: 10.1016/j.xcrm.2024.101586. Epub 2024 May 9. Cell Rep Med. 2024. PMID: 38723625 Free PMC article. No abstract available.
Abstract
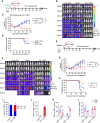
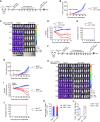
Pediatric patients with relapsed or refractory rhabdomyosarcoma (RMS) have dismal cure rates, and effective therapy is urgently needed. The oncogenic receptor tyrosine kinase fibroblast growth factor receptor 4 (FGFR4) is highly expressed in RMS and lowly expressed in healthy tissues. Here, we describe a second-generation FGFR4-targeting chimeric antigen receptor (CAR), based on an anti-human FGFR4-specific murine monoclonal antibody 3A11, as an adoptive T cell treatment for RMS. The 3A11 CAR T cells induced robust cytokine production and cytotoxicity against RMS cell lines in vitro. In contrast, a panel of healthy human primary cells failed to activate 3A11 CAR T cells, confirming the selectivity of 3A11 CAR T cells against tumors with high FGFR4 expression. Finally, we demonstrate that 3A11 CAR T cells are persistent in vivo and can effectively eliminate RMS tumors in two metastatic and two orthotopic models. Therefore, our study credentials CAR T cell therapy targeting FGFR4 to treat patients with RMS.
Keywords: CAR T cell therapy; FGFR4; rhabdomyosarcoma; specific cytotoxicity.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests J. Khan, R.J.O., D.S.D., and A.T.C. are inventors on international patent application no. PCT/US2016/052496. The 3A11 CAR sequence is in this patent application (see https://patents.justia.com/patent/11078286) filed on September 19, 2016, titled “Monoclonal antibodies specific for fibroblast growth factor receptor 4 (FGFR4) and methods of their use.”
Figures






References
-
- Shern J.F., Chen L., Chmielecki J., Wei J.S., Patidar R., Rosenberg M., Ambrogio L., Auclair D., Wang J., Song Y.K., et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–231. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

