Comparative proximity biotinylation implicates the small GTPase RAB18 in sterol mobilization and biosynthesis
- PMID: 37774976
- PMCID: PMC10641524
- DOI: 10.1016/j.jbc.2023.105295
Comparative proximity biotinylation implicates the small GTPase RAB18 in sterol mobilization and biosynthesis
Abstract
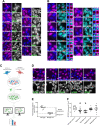
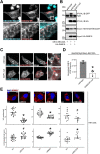
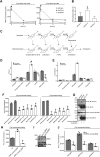
Loss of functional RAB18 causes the autosomal recessive condition Warburg Micro syndrome. To better understand this disease, we used proximity biotinylation to generate an inventory of potential RAB18 effectors. A restricted set of 28 RAB18 interactions were dependent on the binary RAB3GAP1-RAB3GAP2 RAB18-guanine nucleotide exchange factor complex. Twelve of these 28 interactions are supported by prior reports, and we have directly validated novel interactions with SEC22A, TMCO4, and INPP5B. Consistent with a role for RAB18 in regulating membrane contact sites, interactors included groups of microtubule/membrane-remodeling proteins, membrane-tethering and docking proteins, and lipid-modifying/transporting proteins. Two of the putative interactors, EBP and OSBPL2/ORP2, have sterol substrates. EBP is a Δ8-Δ7 sterol isomerase, and ORP2 is a lipid transport protein. This prompted us to investigate a role for RAB18 in cholesterol biosynthesis. We found that the cholesterol precursor and EBP-product lathosterol accumulates in both RAB18-null HeLa cells and RAB3GAP1-null fibroblasts derived from an affected individual. Furthermore, de novo cholesterol biosynthesis is impaired in cells in which RAB18 is absent or dysregulated or in which ORP2 expression is disrupted. Our data demonstrate that guanine nucleotide exchange factor-dependent Rab interactions are highly amenable to interrogation by proximity biotinylation and may suggest that Micro syndrome is a cholesterol biosynthesis disorder.
Keywords: BioID; EBP; ORP2; RAB18; Rab; SNARE proteins; cholesterol metabolism; lathosterol; lipid transport; protein–protein interaction.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Zhen Y., Stenmark H. Cellular functions of Rab GTPases at a glance. J. Cell Sci. 2015;128:3171–3176. - PubMed
-
- Lamber E.P., Siedenburg A.-C., Barr F.A. Rab regulation by GEFs and GAPs during membrane traffic. Curr. Opin. Cell Biol. 2019;59:34–39. - PubMed
-
- Aligianis I.A., Johnson C.A., Gissen P., Chen D., Hampshire D., Hoffmann K., et al. Mutations of the catalytic subunit of RAB3GAP cause Warburg Micro syndrome. Nat. Genet. 2005;37:221–223. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

