Baseline Expression of Immune Gene Modules in Blood is Associated With Primary Response to Anti-TNF Therapy in Crohn's Disease Patients
- PMID: 37776235
- PMCID: PMC10906954
- DOI: 10.1093/ecco-jcc/jjad166
Baseline Expression of Immune Gene Modules in Blood is Associated With Primary Response to Anti-TNF Therapy in Crohn's Disease Patients
Abstract
Background and aims: Anti-tumour necrosis factor [anti-TNF] therapy is widely used for the treatment of inflammatory bowel disease, yet many patients are primary non-responders, failing to respond to induction therapy. We aimed to identify blood gene expression differences between primary responders and primary non-responders to anti-TNF monoclonal antibodies [infliximab and adalimumab], and to predict response status from blood gene expression and clinical data.
Methods: The Personalised Anti-TNF Therapy in Crohn's Disease [PANTS] study is a UK-wide prospective observational cohort study of anti-TNF therapy outcome in anti-TNF-naive Crohn's disease patients [ClinicalTrials.gov identifier: NCT03088449]. Blood gene expression in 324 unique patients was measured by RNA-sequencing at baseline [week 0], and at weeks 14, 30, and 54 after treatment initiation [total sample size = 814].
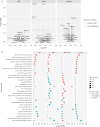
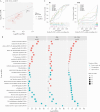
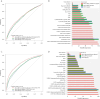
Results: After adjusting for clinical covariates and estimated blood cell composition, baseline expression of major histocompatibility complex, antigen presentation, myeloid cell enriched receptor, and other innate immune gene modules was significantly higher in anti-TNF responders vs non-responders. Expression changes from baseline to week 14 were generally of consistent direction but greater magnitude [i.e. amplified] in responders, but interferon-related genes were upregulated uniquely in non-responders. Expression differences between responders and non-responders observed at week 14 were maintained at weeks 30 and 54. Prediction of response status from baseline clinical data, cell composition, and module expression was poor.
Conclusions: Baseline gene module expression was associated with primary response to anti-TNF therapy in PANTS patients. However, these baseline expression differences did not predict response with sufficient sensitivity for clinical use.
Keywords: Anti-TNF; Crohn’s disease; transcriptomic biomarkers.
© The Author(s) 2023. Published by Oxford University Press on behalf of European Crohn’s and Colitis Organisation.
Conflict of interest statement
Mark Reppell, Nizar Smaoui, Jeffrey F. Waring, Valerie Pivorunas, and Heath Guay are employees of AbbVie and may own stock and/or options. Simeng Lin reports non-financial support from Pfizer outside the submitted work. James R. Goodhand reports grants from F. Hoffmann-La Roche AG, grants from Biogen Inc, grants from Celltrion Healthcare, grants from Galapagos NV, and non-financial support from Immundiagnostik outside the conduct of the study. Nicholas A. Kennedy reports grants from F. Hoffmann-La Roche AG, grants from Biogen Inc, grants from Celltrion Healthcare, grants from Galapagos NV, and non-financial support from Immundiagnostik; grants and non-financial support from AbbVie, grants and personal fees from Celltrion, personal fees and non-financial support from Janssen, personal fees from Takeda, and personal fees and non-financial support from Dr Falk, outside the submitted work. Tariq Ahmad reports grants and non-financial support from F. Hoffmann-La Roche AG, grants from Biogen Inc, grants from Celltrion Healthcare, grants from Galapagos NV, and non-financial support from Immundiagnostik; personal fees from Biogen Inc, grants and personal fees from Celltrion Healthcare, personal fees and non-financial support from Immundiagnostik, personal fees from Takeda, personal fees from ARENA, personal fees from Gilead, personal fees from Adcock Ingram Healthcare, personal fees from Pfizer, personal fees from Genentech, and non-financial support from Tillotts, outside the submitted work. Carl A. Anderson has received consultancy or lectureship fees from Genomics plc, BridgeBio, and GSK. The remaining authors have no conflicts of interest to report.
Figures