Anticodon sequence determines the impact of mistranslating tRNAAla variants
- PMID: 37776539
- PMCID: PMC10543346
- DOI: 10.1080/15476286.2023.2257471
Anticodon sequence determines the impact of mistranslating tRNAAla variants
Abstract
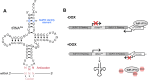
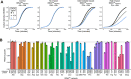
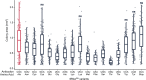
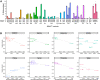
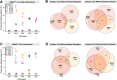
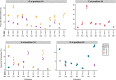
Transfer RNAs (tRNAs) maintain translation fidelity through accurate charging by their cognate aminoacyl-tRNA synthetase and codon:anticodon base pairing with the mRNA at the ribosome. Mistranslation occurs when an amino acid not specified by the genetic message is incorporated into proteins and has applications in biotechnology, therapeutics and is relevant to disease. Since the alanyl-tRNA synthetase uniquely recognizes a G3:U70 base pair in tRNAAla and the anticodon plays no role in charging, tRNAAla variants with anticodon mutations have the potential to mis-incorporate alanine. Here, we characterize the impact of the 60 non-alanine tRNAAla anticodon variants on the growth of Saccharomyces cerevisiae. Overall, 36 tRNAAla anticodon variants decreased growth in single- or multi-copy. Mass spectrometry analysis of the cellular proteome revealed that 52 of 57 anticodon variants, not decoding alanine or stop codons, induced mistranslation when on single-copy plasmids. Variants with G/C-rich anticodons resulted in larger growth deficits than A/U-rich variants. In most instances, synonymous anticodon variants impact growth differently, with anticodons containing U at base 34 being the least impactful. For anticodons generating the same amino acid substitution, reduced growth generally correlated with the abundance of detected mistranslation events. Differences in decoding specificity, even between synonymous anticodons, resulted in each tRNAAla variant mistranslating unique sets of peptides and proteins. We suggest that these differences in decoding specificity are also important in determining the impact of tRNAAla anticodon variants.
Keywords: Mistranslation; expanded decoding; genetic code; tRNA biology; tRNAAla.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
